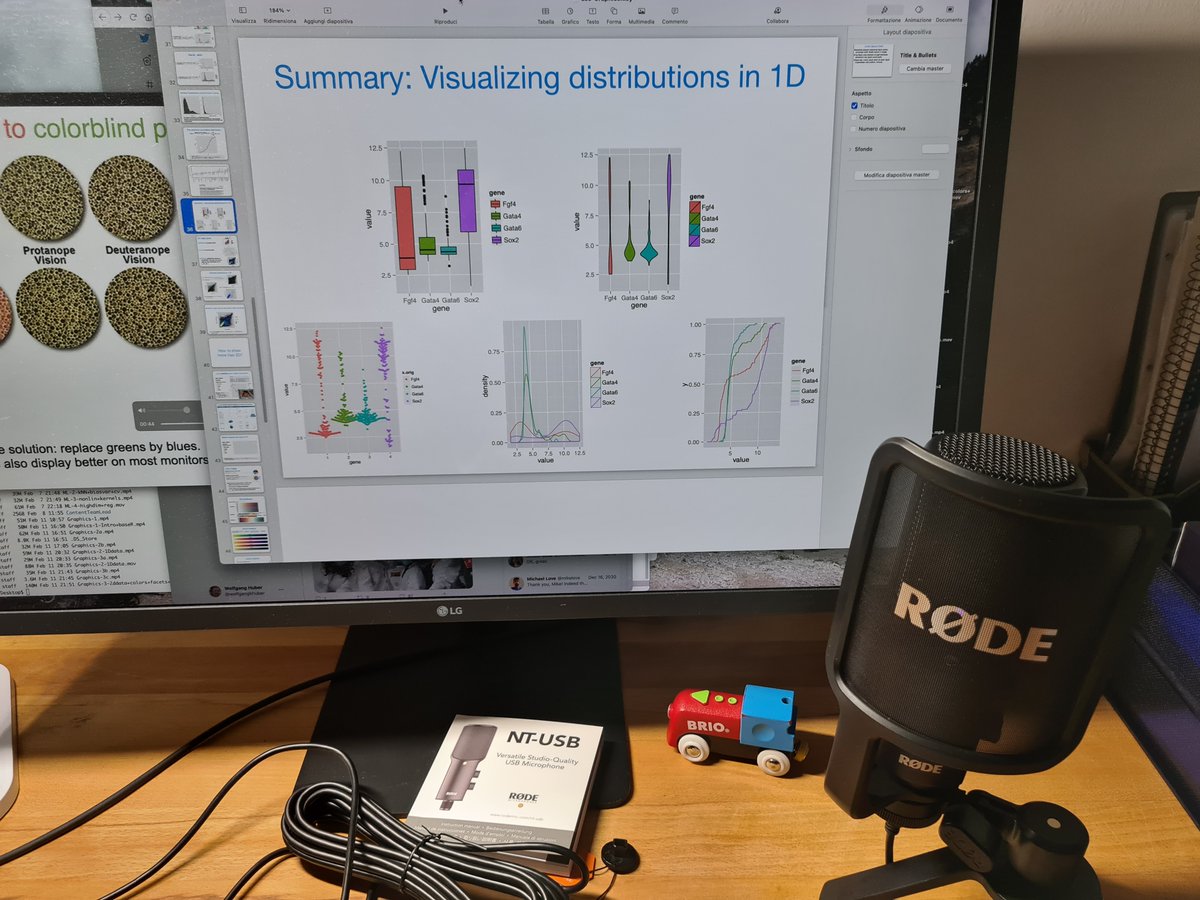
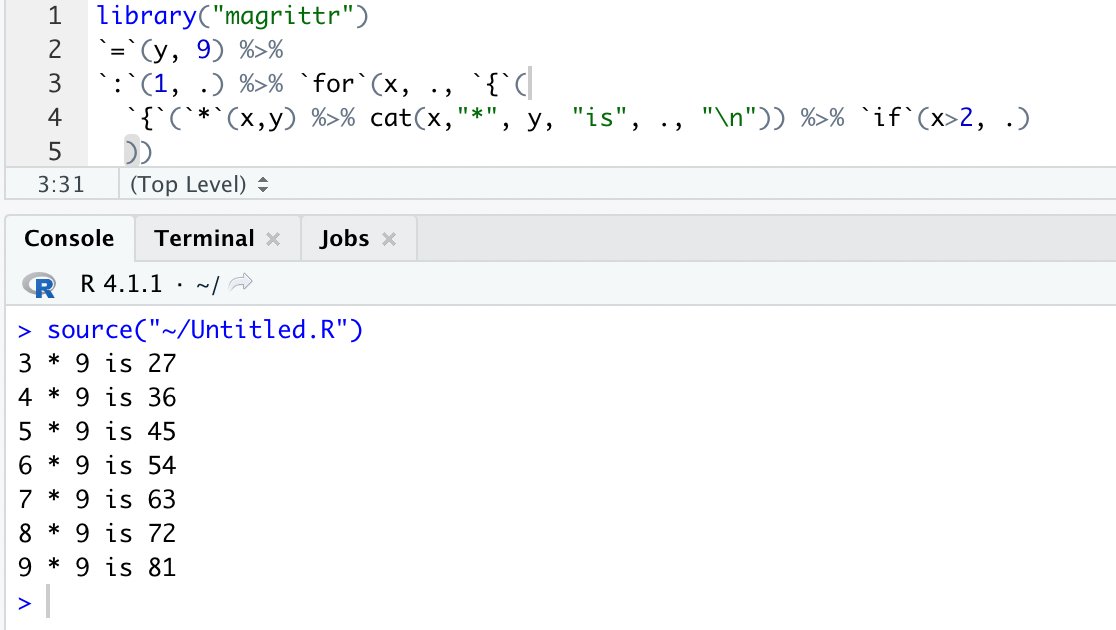
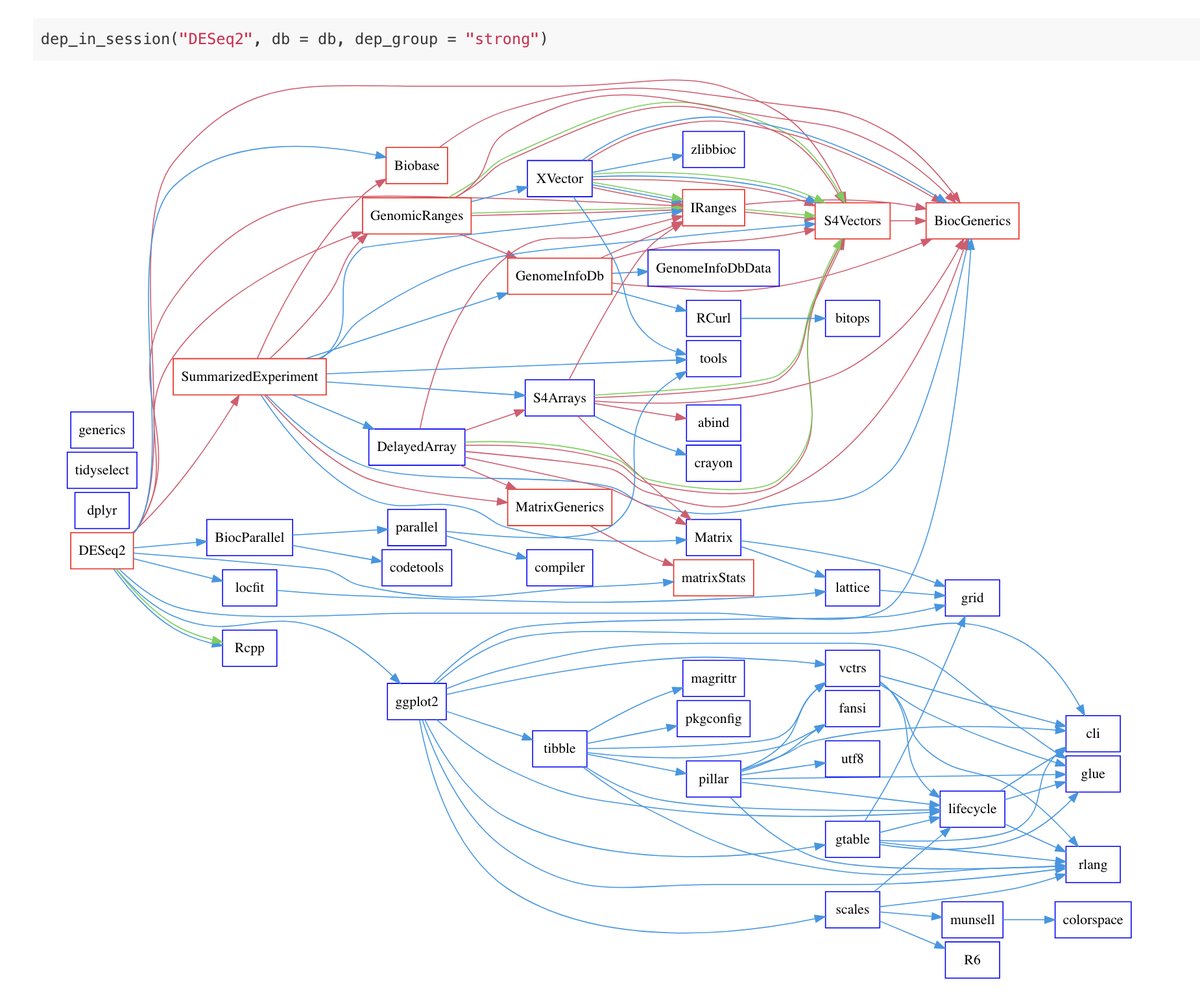
library("jsonlite")
library("dplyr")
library("stringr")Repository with the source code: https://github.com/wolfganghuber/tweets
The file data/tweets.js is in the Twitter archive (zip file) that I downloaded from X. Adapt path to whatever you have.
archivepath = "/Users/whuber/twitter/data"
tweets = readLines(file.path(archivepath, "tweets.js")) |>
sub("^window.YTD.tweets.part0 = ", "", x = _) |>
fromJSON(flatten = TRUE) Select tweets and select relevant columns. Here, I chose to drop all retweets and keep all others. Adapt this to your liking.
isrt = grepl("^RT", tweets$tweet.full_text)
out = dplyr::select(tweets[!isrt, ], all_of(c(
date = "tweet.created_at",
text = "tweet.full_text",
id = "tweet.id",
retweets = "tweet.retweet_count",
likes = "tweet.favorite_count",
mediadf = "tweet.entities.media")
))Some cleanup and prettification: add hyperlinks to URLs and tweet IDs, and sort by date (default: ascending).
out = mutate(out,
text = str_replace_all(text, "(https?://\\S+)", "<a href='\\1'>\\1</a>"),
idhtml = sprintf('<a href="https://x.com/wolfgangkhuber/status/%s">%s</a>', id, id),
date = strptime(out$date, "%a %b %d %H:%M:%S %z %Y", tz = "UTC")
) |> arrange(date)Deal with media. Tweets that have media associated (images, movies) come with a data.frame in the tweet.entities.media column. We also just go and find all media whose filename contains the tweet ID (see code for ip below) and check consistency.
indir = file.path(archivepath, "tweets_media")
outdir = "media"
mediafiles = dir(indir)
out$media = character(nrow(out))
if (file.exists(outdir))
unlink(outdir, recursive = TRUE)
dir.create(outdir)
for (i in seq_len(nrow(out))) {
m = out$mediadf[[i]]
if (!is.null(m)) {
stopifnot(is.data.frame(m), nrow(m) == 1)
key = tools::file_path_sans_ext(basename(m$media_url))
im = grep(key, mediafiles)
ip = grep(paste0("^", out$id[i]), mediafiles)
if (length(ip) == 0) {
message(sprintf("%s from tweet #%d not found", key, i))
} else {
stopifnot(im %in% ip)
file.copy(file.path(indir, mediafiles[ip]), outdir)
out$media[i] = paste(
"::: {.tweet-media}",
paste(sprintf('{.lightbox .resized-image}', file.path(outdir, mediafiles[ip])), collapse = "\n"),
":::", sep = "\n")
}
}
}Create the markdown text for each tweet. The main work here is done by the CSS file.
tweetsmd = with(out, sprintf(
'::: {#%s .tweet}
::: {.tweet-header}
<span class="tweet-timestamp">%s Retweets: %s Likes: %s</span>
<span class="tweet-handle">%s</span>
:::
::: {.tweet-content}
%s
:::
%s
:::
', id, as.character(date), retweets, likes, idhtml, text, media
))Inject into the document.
cat(tweetsmd, "\n", sep = "")Final meeting RADIANT EU project: bring statistics to the masses. Produced edgeR-robust,DESEq2,HTSeq,flipflop,INSPEcT http://t.co/AfDhaEiiAh
IONiseR to visualise Nanopore data and see batch effects http://t.co/Y1pn6gMlLR and BitSeq - also from RADIANT
@JimJohnsonSci @Y_Gilad @mbeisen @HarmitMalik @bdelloid Recommendation of papers can be post-pub, incl by professionals; think films, books
@HarmitMalik @mbeisen @bdelloid Problem is not type of review (maybe want pre and post) but its gate keeper function -obsolete with internet
R Journal new issue: http://t.co/EddyfTapbB - new packages, graphics
@genetics_blog Good points -but misinformed about high level languages and big data. Bad workman blames his tools.
Bioconductor annual conference - connect software and biology. July 20-22 in Seattle http://t.co/J7uGIGVMTd
@genetics_blog Good points: need more professional developers, long-term software maintenance, service attitude,funding,academic recognition
Bioconductor channel in F1000Research - for cross-package end-to-end solution workflows http://t.co/a6JxOm7rK5
@lpachter R session with 40 packages loaded, all from different authors. Who says no one builds on others’ code?
@lpachter Building = using building blocks to create something bigger. Many workflows, >20 packages depend on DESeq/2 http://t.co/qbv1dbbKq4
@lpachter Free for non-commercial is a legal muddle, impractical (eg private unis) Subsidies flow 2 ways, commerce pays taxes pay academics.
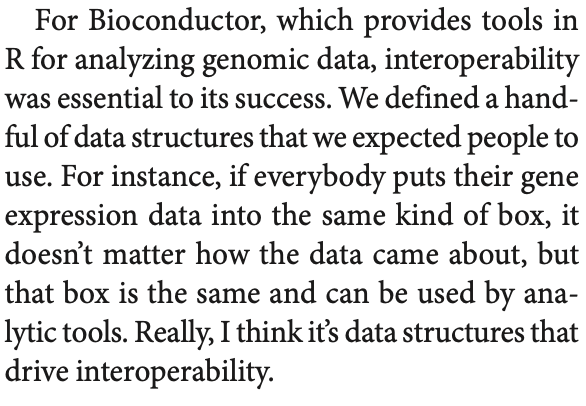
@lpachter Dozens, incl open source packages it relies on for stats, graphics, I/O. It’s a building, not a monolith. Interoperability rocks.
@lpachter Not proposing writing by comittee- each package is best written by one person. Figuring that out is key http://t.co/bkRH8I4D75
Computational postdocs at EMBL and NCT in high-throughput phenotyping & multiomics for precision oncology http://t.co/RxYLSUFMIU
EMBL Interdisciplinary postdoc fellowships - http://t.co/dQMwYiNOKH
EMBL-Stanford conference on (gen)omics and health 16-19 Nov, abstract submission is open http://t.co/0I5FsrurQH
S3/++ Summer School of Science for high-school students in Croatia http://t.co/2Rbw8BNErs
@ewanbirney what kind of anatomical features would make exp( i ) = -1 more intuitive
@ewanbirney Yes, the hill is the best part of it: cardio in the morning, really quick ride home in the evening, either asphalt or bumpy
Complete analysis as a knitr vignette in Suppl. File 2. https://t.co/TW3QshC22M
@JinliangYang @xieyihui R package containing data & vignette is submitted to Bioconductor, in the meanwhile http://t.co/Vxa2sZSRSw
Could Greece become prosperous again? John PA Ioannidis http://t.co/6Hy3ooUwLy
Beach read: The Big Short by Michael Lewis, on subprime crisis of ’00s, cynical Wall St wizkids, complicit rating agencies, inept regulators
Wonder whether there is an equivalent of shorting (a bond, a stock) in the world of science?
HilbertViz by S.Anders came with #Bioconductor 2.3, 10/2008. New HilbertCurve by Zuguang Gu, more flexible interface https://t.co/UjRCXdctYi
On non-standard evaluation in R and Python, and its use in data analysis https://t.co/zDkdfYsYtr
Enjoying Feynman’s “Surely You’re Joking”. Should have done this long ago already. https://t.co/s1DB6FArnz!
Abstract submission C1omics “Single-cell Omics methods & applicns” in Manchester (Nov 24-25) is open, deadline Sep 14 http://t.co/GG508KqpTa
Microbiome can predict preterm birth: http://t.co/v1b1RUPUZf Fully reproducible data analysis: http://t.co/kscbVpW0FR
@casey6r0wn Fig. 3 and 4B. (And yes they acknowledge that sample size is limiting and more data needed.)
@casey6r0wn ‘predict’ here really means that microbiome state temporally precedes preterm birth. Not the statistics/ML abuse of the word.
@casey6r0wn @SherlockpHolmes yes the semantics are important. praedicere: prae ‘beforehand’ + dicere ‘say’. No causality, but temp. order.
The Case for Teaching Ignorance http://t.co/0LdwS37p18
Support for all bioC packages in the forum: https://t.co/TZKnZijB0k biomaRt pkg is an interface, has no “content”. https://t.co/Rg98EngFkS
@rafalab @stat110 Insurance? Also highlights independence assumption.
@rafalab @stat110 Going to the beach, and skiing? I.e. Liquid-gas and solid-liquid phase transitions.
Interdisciplinary Postdoc Fellowships: deadline Sep 10, discuss projects with PIs before http://t.co/RZRalxodh6 #EMBL http://t.co/osr3yNuDjx
Differential Splice Junction Usage and Intron Retentions with DEXSeq https://t.co/LU7Yq6DFWG
@hadleywickham As far as I understand my language, the 24h clock is used and 10:10 always implies AM, otherwise you write 22:10.
Drug-protein target identification by Thermal Proteome Profiling - methods paper incl. @Bioconductor software http://t.co/6lfbty91gP
@Bioconductor Developer Meeting,7-8 Dec 2015, Cambridge UK. For regulars and newbies https://t.co/lgXLLDU2Sg http://t.co/y6URTpChm2
C1omics: single-cell omics methods and applications CRUK Manchester 24-25 Nov http://t.co/GG508KqpTa http://t.co/Ux4pjClbu7
Never be afraid to try something new. Remember, amateurs built the ark. Professionals built the Titanic.
Love BSD #fortune
Jupyter notebook is also available https://t.co/Kli0Abc1qU in Docker container https://t.co/rJna3XR6Is https://t.co/EZsMrMJPgn
Yes. And Vlad Kim will be posting a video explaining the process. https://t.co/SfC9SLR2Oe
Hadn’t realized how few journals have a clear policy on correcting papers once out. @embojournal now does http://t.co/rlGdECsfnF
Postdoc - data science in multi-omics and personalized medicine https://t.co/PGNfnuBoWc https://t.co/IQsFSuzbsh
@genetics_blog @stevepiccolo Is this not first a question of semantics and only then of statistics, experiment design? what is ‘expressed’?
Cancerletter article by Baggerly and Gunsalus on the ORI investigation of Potti/Duke case https://t.co/87D9kuGVi9
R debugging and robust programming course by Laurent Gatto & Robert Stojnic at @EMBLorg #rstats https://t.co/hoKykfzZPM
TimerQuant: Modelling tandem fluorescent timer design and data for measuring protein turnover in embryos https://t.co/YZaUYOg40c
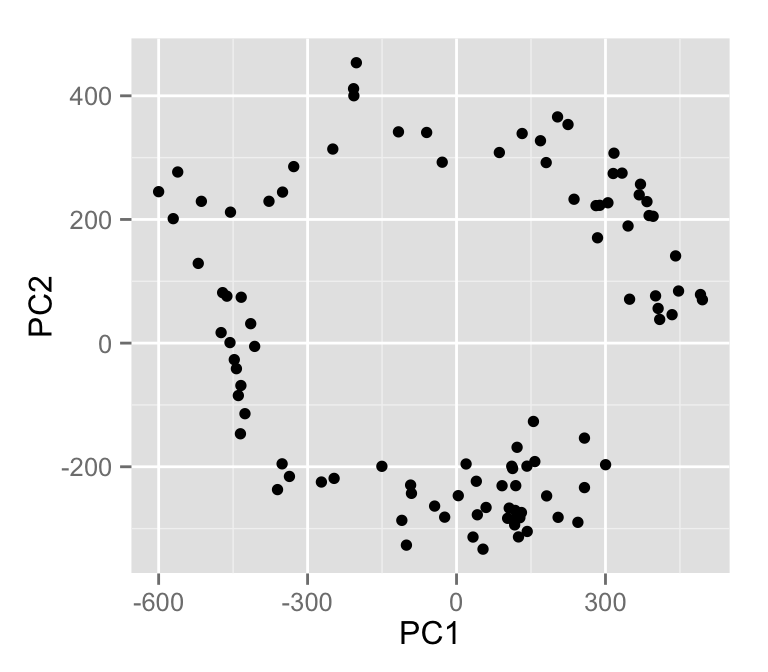
Cycles and horseshoes in PCA plots - even if there is nothing curvilinear in the data https://t.co/5UydcreBwf https://t.co/VuqNN3VPj0
@JennyBryan They just know how good you are.
Also available as Rmarkdown executable document https://t.co/imCJCd6k8x and Jupyter notebook https://t.co/rJna3XR6Is https://t.co/BAyg8brOD8
A publications list boiled down to what really matters. Brilliant. https://t.co/x0cd7k9jtp https://t.co/nIyZiycnM5
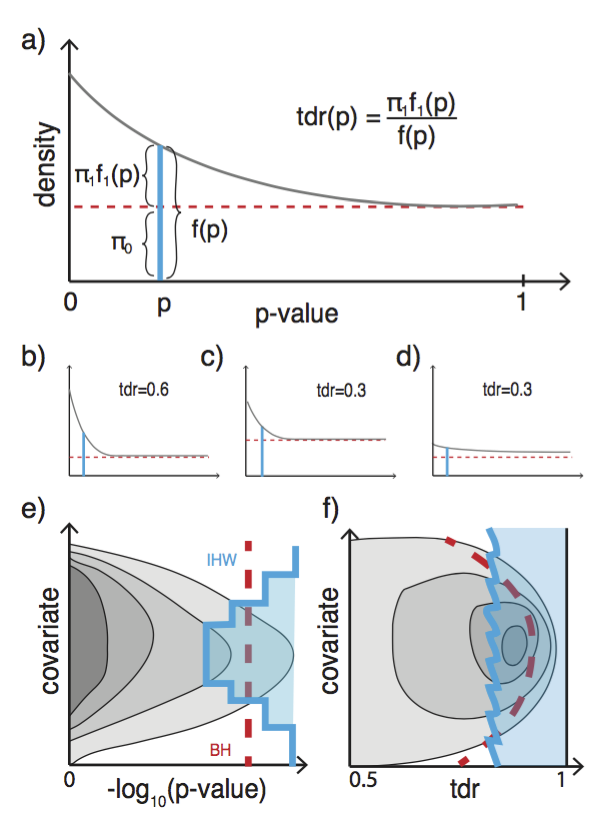
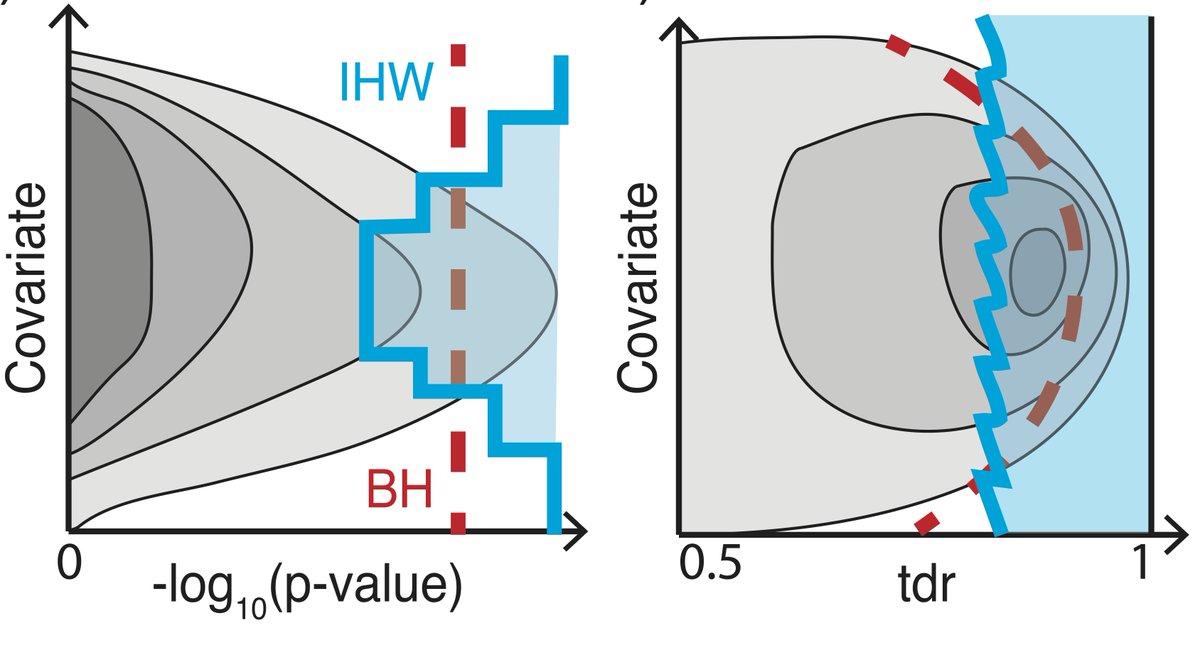
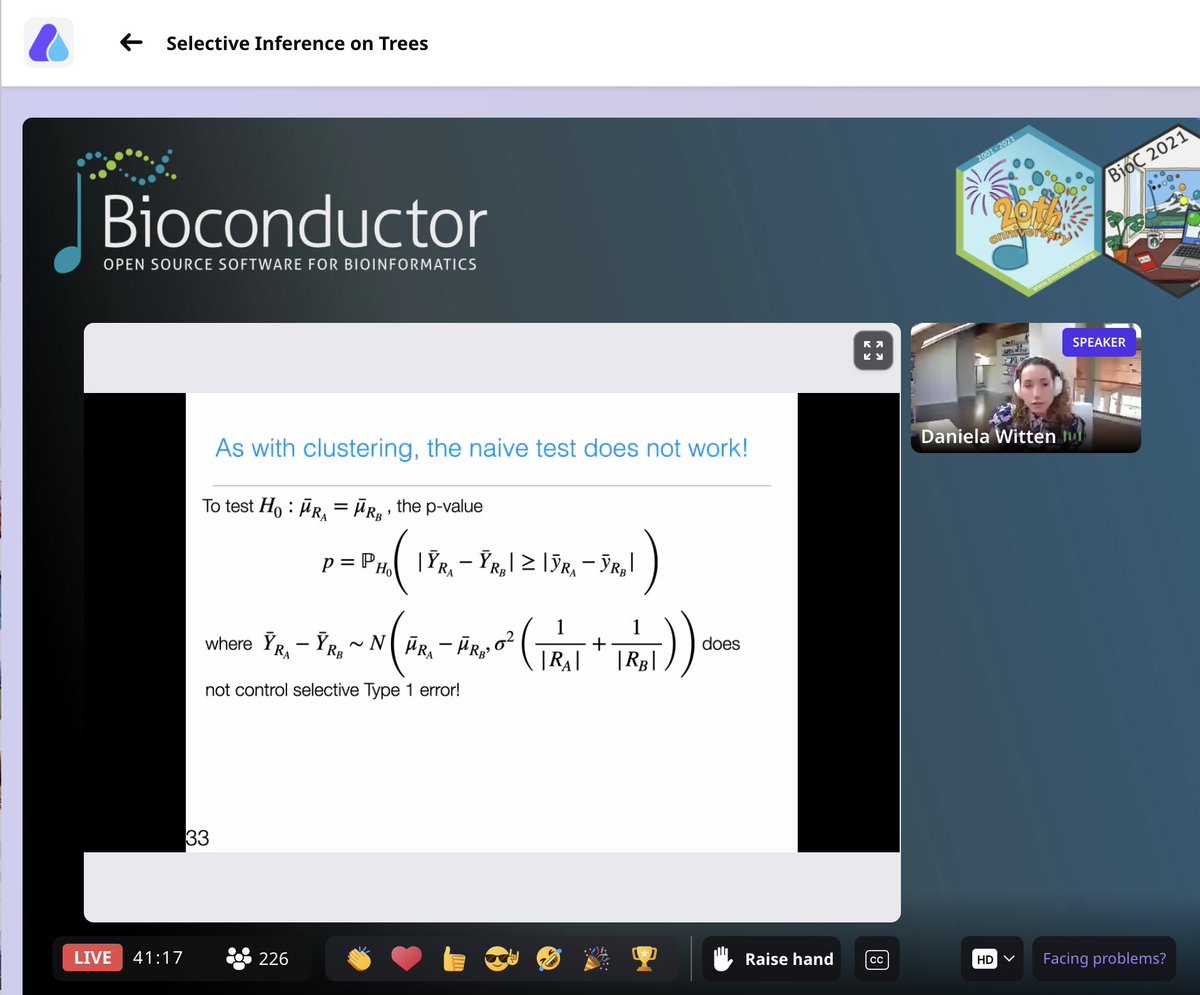
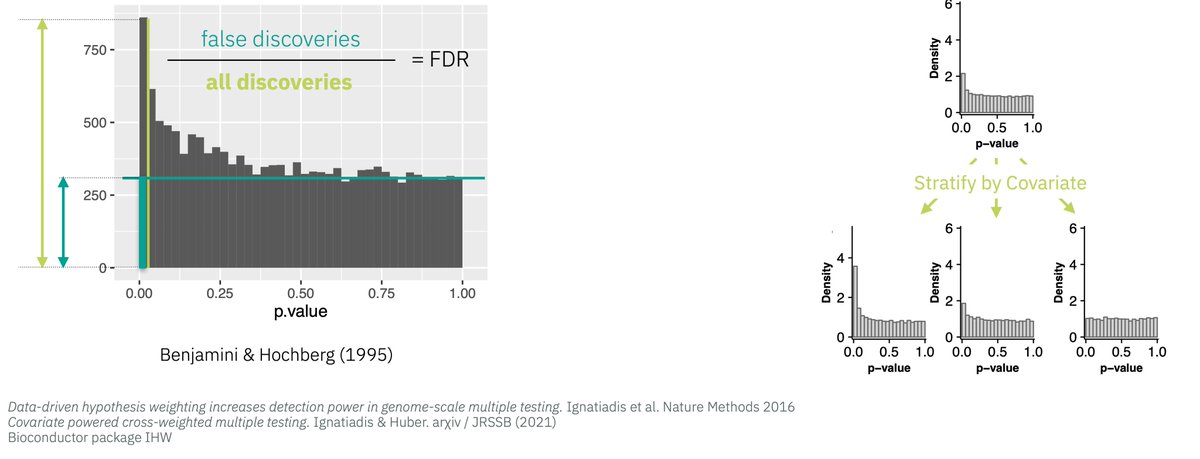
Data-driven hypothesis weighting in multiple testing- RNASeq, GWAS, proteomics etc, bioRχiv https://t.co/61esOg3Gq6 https://t.co/t6aZKhghkl
@jtleek @mikelove Correct. And if a method’s type-I error control is not independent of covariate, it’s poorly calibrated & can be improved.
@jtleek @mikelove They’re abundant: RNA-Seq - avg. counts; eQTL - distance, TADs; proteomics - no. of peptides; neuron pairs - distance; etc
Statistical Computing workshop July 17-20, 2016 on Schloß Reisensburg - call for contributions https://t.co/ehPkZpynxC
Gene-level vs transcript-level differential expression dissected -by data analysis rather than made-up toy examples https://t.co/kHEVc5Xrem
Bioconductor moving from closed pre-publication review model to open post-publication peer review https://t.co/7vUetQGnii
New issue of the #rstats journal https://t.co/o03DYPwBpV
Genomic Medicine in Lymphoma - CancerCoreEurope conference https://t.co/RKHWByuIFF https://t.co/mMb0Llqw9G
@MagnusRattray @mikelove It will only be complete with Love.
The spreading of misinformation online - conspiracy theorists, scientists, echo chambers: a data analysis https://t.co/PoH9M8hF6n
@ewanbirney library(ggmap) library(geosphere) p=c(“BOS”,“LHR”,“TLV”) x=geocode(p) distGeo(x[1,],x[2,]) distGeo(x[2,],x[3,]) #5255 v 3593 km
@ewanbirney @timtriche good to think of Britain close to Europe and not in the middle of the Atlantic
The Trojan Wars of Machine Learning https://t.co/1M0GKdZqW7
Artur Fischer: inventor, businessman and creator of construction toys https://t.co/T3OhB4thz4 https://t.co/lBEYrkEzAx
Exciting postdoc opportunity with dual research / building infrastructures (→ELIXIR) slant https://t.co/KBiTwMX4QA https://t.co/asAkoZwuaD
EMBL Advanced Course: R Debugging and Robust Programming -with L Gatto & R Stojnic 25-26 Feb https://t.co/hoKykfzZPM https://t.co/aGrFCzVNUC
@StephenEglen Had trouble with rpubs to update, needed new URL: https://t.co/h58CM9jU0c ‘Cycles and horseshoes in PCA plots’ - weird indeed
@StephenEglen @lgatto Mine. Many ESS users out there. Recently I prefer RStudio for Rmarkdown, R-shell integration, debugging & fewer bugs.
@StephenEglen Great find! Yes, these researchers pretty much nailed it in 1995. If only they had already had R/knitr then.
Time from paper submission to publication is an anachronism https://t.co/nnNfb6OzYT
@ewanbirney Symmetry groups of dodecahedron & icosahedron are same, eg. https://t.co/BEo4e1IMK5, maybe it’s complexity (no. vertices, edges)
@ewanbirney Shape of most macroscopic objects has little to do with molecular orbitals
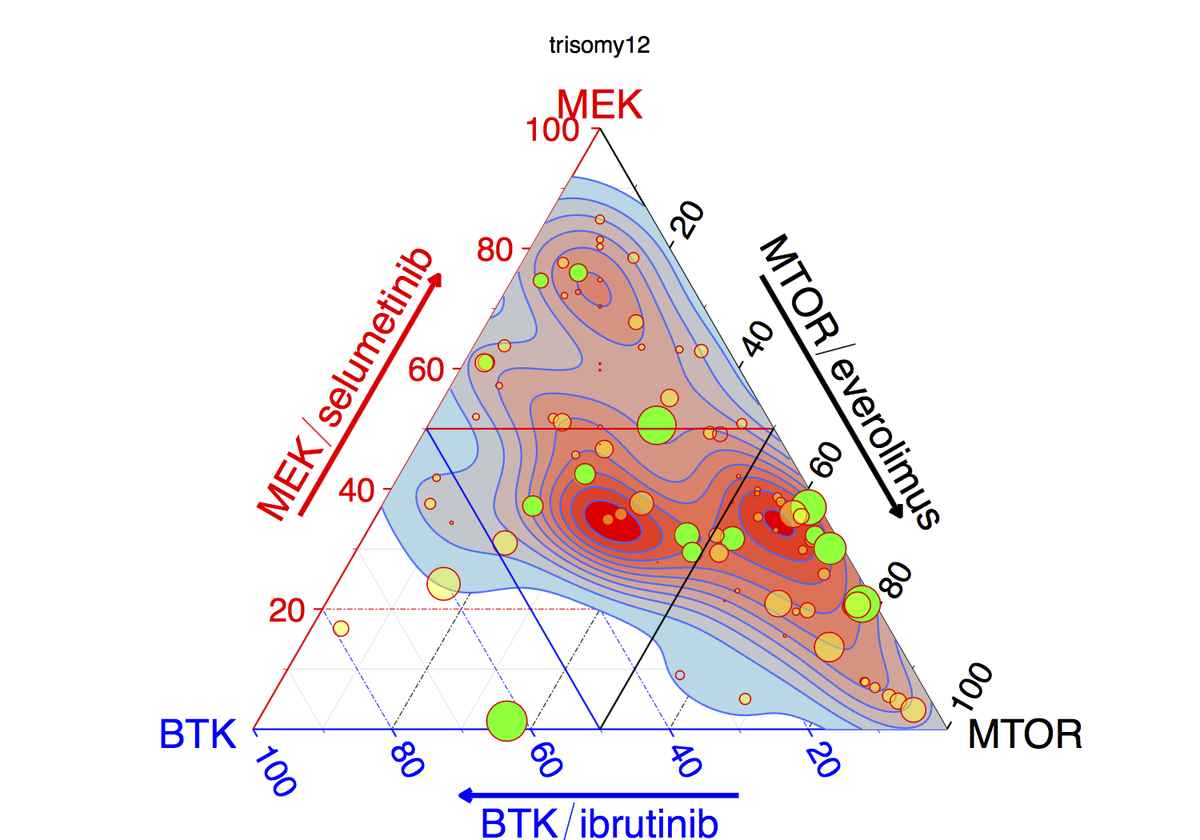
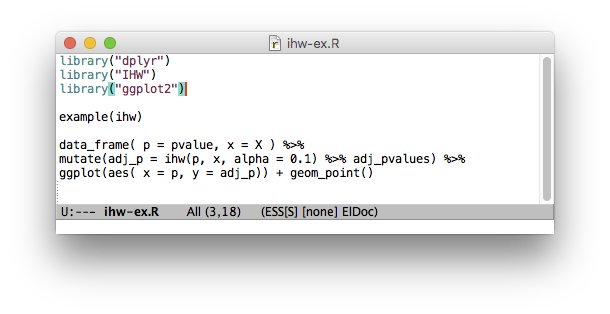
Now on #Bioconductor: IHW for FDR with data-driven hypothesis weighting https://t.co/dUP9D4tGjf preprint is here: https://t.co/61esOg3Gq6
Exciting postdoc opportunities w Misha Savitski @EMBL - protein chemistry, chemical biology https://t.co/MrzHiIQ4bB https://t.co/0OojkOGKFp
@noort_zuit @ClaessenLAB Much has changed in 10 yrs. For Bioconductor, good place to start: https://t.co/F9Q7EZSsOj https://t.co/FKEgNwBWLO
@mbeisen short, polite and informative comment in @PubMedCommons ?
Underpowered me-too experiment #4wordacademicsadstory
Good news: towards sustainable bioinformatics infrastructures through international organization https://t.co/69CzWSlQNf
These were the coolest construction sets - Artur Fischer obituary in NYT https://t.co/M9hJOK2rgO https://t.co/qjprAos0VH
Increased science funding goes primarily into administrators, not scientists: https://t.co/r7lQwOt1nz (in German)
“Our job is to make sure you don’t do anything wrong”- admin director of ‘excellent’ German uni to colleague who took over a chair&institute
Conference - Systematic Functional Annotation of Cancer Variants. Heidelberg 19-21 May https://t.co/U2zYwIPClZ https://t.co/sNKFA7Ediq
@markrobinsonca @CSoneson If covariate not involved in dependence, same as BH. If yes, gets complex: criterion for choice @nikosIgnatiadis
@jkpritch https://t.co/R05CTs7I0V
@michaelhoffman @BioMickWatson @talyarkoni <-(“x”, pi) library(“magrittr”) pi %>% <-(“x”)
pi %>% names<-(“pi”) -> x
Week-long R/@Bioconductor course “Statistics and Computing in Genome Data Science” in Brixen https://t.co/EU8AbbhpM7 https://t.co/84GgxYRbXp
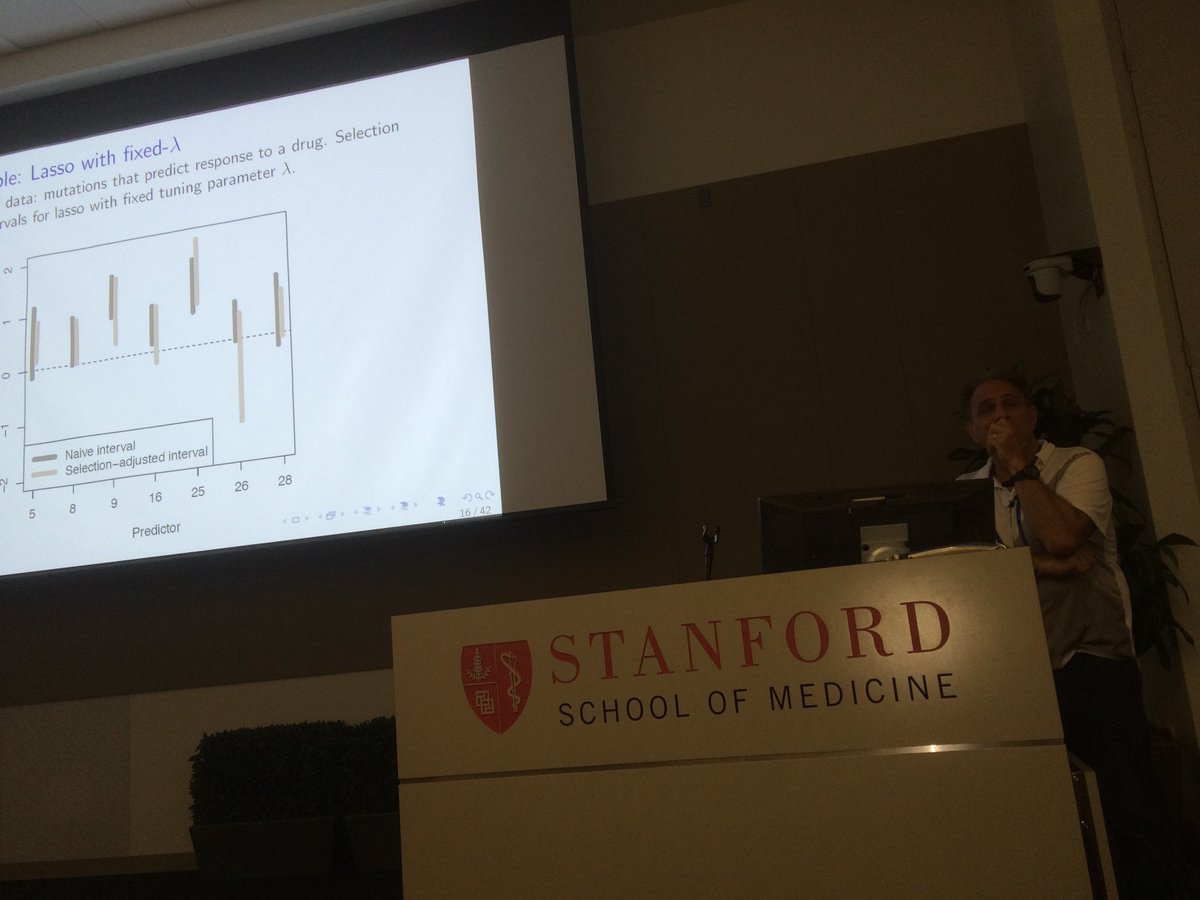
Have you been told you can’t “snoop” data for model selection, then still do valid inference (p-values)? Now you can https://t.co/h5rxPoVipv
@psychgenomics @JennyBryan @michaelhoffman @michelebusby one thing though: log is dimension-agnostic, log(ax)=log(x) up to constant (1/2)
@psychgenomics @JennyBryan @michaelhoffman @michelebusby asinh is not, choice of scale matters (but is often implied by application) (2/2)
Statistical causal inference in genetics - Montreal July 25-29, 2016 https://t.co/5WnrZbxYcO
Command line weather report #wttr.in curl https://t.co/tsOhVLZQHH https://t.co/gZYGBWKFUT
Your paper about a Bioc workflow, package or teaching lab: call for papers https://t.co/FKEgNwBWLO https://t.co/TKcQr5WGCR
@JennyBryan @rdpeng lapply(1:n, function(i) lapply(1:m, function(j) {…} )%>% do.call(rbind, .) )%>% do.call(rbind, .) is your friend
Thank you @nickschurch @mikelove @gjbarton for bringing this to light. Great dataset & study design. What about Fig.3 (tool similarity)?
@biobenkj Thank you. Perhaps an unintended consequence of system maintenance last week. We’ll get this fixed asap @andrzejkoles
@biobenkj @andrzejkoles All back to normal. We had suffered a brute-force password attack that caused DoS. Our EMBL IT team fixed it.
@jgschraiber Provide raw data & fully document analysis (eg knitr, Jupyter) so others can vary params & check robustness
Clinical Bioinformatics as a Service - Workshop at ECCB 2016 Announcement & call for papers: https://t.co/gpS5FKbHnt
@jaspattwit @BioMickWatson Submit talk proposal via website.
(‘Call for papers’ in prev. tweet really meant ‘Call for contributed talks’.)
Industry Postdoc @EMBL: Statistical Computing in Multi-Omics and Drug Target Validation https://t.co/r2VwFYq10S https://t.co/mPIZATvHd4
David Miliband: Britain has for centuries been a firefighter. This is no time for it to join the ranks of arsonists https://t.co/J9BpRX1J7N
Political prestige project https://t.co/dD0L4qA9UZ
Thanks to @gjbarton @nickschurch @mikelove for highly professional resolution. A landmark dataset and paper!
@mikelove @nomad421 @nikosIgnatiadis Indeed with IHW, hypotheses with low weights (‘filtered’ in Independent Filtering) do get an adj.p of 1
Peer review: the imagined functions of this institution are in flux, but they were never as fixed as many believe. https://t.co/sInlnQGA2J
Cancer Systems Genetics Conference 19-21 May in Heidelberg https://t.co/HwJ2f1OOe8 Registration closing soon! https://t.co/DpgkutQCk0
NSF test finds eliminating deadlines halves number of grant proposals - and makes them better https://t.co/8nz1Kcs1ND
Just discovered Wikipedia also does Alemannisch: https://t.co/O9Otu577uq
@tuuliel @mattmeier @drchriscole @BioMickWatson Not a panacea. Since ranks ignore signal strength, may be more susceptible to batch effects.
High-energy physics (HEP) software foundation -coordination & common efforts in HEP software https://t.co/rBDiNOQvwa Workshop Paris 2-4 May
Blessed is who, having nothing to say, abstains from giving wordy evidence of the fact. George Eliot
Bioconductor Release 3.3: many new great software packages, 1211 in total. https://t.co/FiODLd26j0
Aim 1 of this grant proposal (paraphrased): “To mine databases and read the literature in the field.” Hmm.
@ewanbirney Enter the Hadleyverse https://t.co/0Fnyix2HXe
@ewanbirney though for SNPs, matrices are the right structure, not dataframes. (& there’s out-of-memory or compressed represntn’s if needed)
Call for contributed talks& flash talks at workshop “Clinical Bioinformatics as a Service” ECCB Den Haag Sep 4, 2015 https://t.co/gpS5FKbHnt
Short MTB break during our quadrennial research review at @EMBL Heidelberg https://t.co/j4AsshT0zg
Cancer Systems Genetics Conference: Shantanu Singh from @DrAnneCarpenter lab on ‘functionalising’ lung cancer mutations with cell painting
Brenda Andrews’ impressive body of work on Genetic Networks: need them for cancer therapies, genotype-phenotype maps https://t.co/fyKHO7CgFa
@dgmacarthur @cazencott And yet it’s not primarily a CS problem. Need orders of magnitude more data- which can only come from model systems.
@thebyrdlab Au contraire. Experiments need to be systematic & large, then are only useful if they result in easily downloadable datasets.
@thebyrdlab Yes. more data needed for variant annotation- but existing data are underanalysed. Better not play off one against the other.
Metagenomics by Illumina amplicon-seq at nucleotide resolution: DADA2 @Bioconductor package doi:10.1038/nmeth.3869 https://t.co/BRIOwXlonS
@HankGreelyLSJU @CaulfieldTim Many journeys would never be made if people knew before how long they are.
@DrAnneCarpenter ..remember there were perfectly functional word processors in 1990s, pre-MSOffice. Progress is not linear, monopolies hurt.
@kwbroman In R #rstats, as in Lisp, code is just data and data can become code. #usedplyr #data_frame https://t.co/5mzzv9ihpI
Beyond ranking by p-value: use additional information in mult. testing, rank by local fdr. https://t.co/fiAM1ejjt5 https://t.co/3h0tH6K3p2
IHW paper BioRxiv https://t.co/61esOg3Gq6 @Bioconductor https://t.co/0c7Zh7zvEV Respect hypothesis diversity,don’t weight them all the same
Switzerland opens the Gotthard base tunnel. Amazing engineering and society achievement. https://t.co/ZSh1rZ90FG
@moorejh @genetics_blog Now out after peer review: https://t.co/fiAM1ejjt5 Don’t rank by p-value only, use additional co-data. #dontwaste
1-week intensive course: Statistical Data Analysis for Genome Biology https://t.co/FaAEvoz5GN Italian Alps July 10-15 Registr’n closing soon
Photoelectric effect and the quantisation of light https://t.co/MZgLJ8FYox
Interested in working for Bioconductor? They’re looking for a web/sys admin allrounder https://t.co/HlqDOrbDiV
Sad. The world just got a lot worse. https://t.co/bSOOfgCXLO
The political categories are no longer “left” and “right”, they’re “open” vs “closed”.
When a close relative did something stupid that will hurt them and the whole family.
@Maxi_Macki @lawrennd EU is a global player and attractive place to live. Brought people together whose grandparents killed each other.
@SherlockpHolmes at #bioc2016 on metagenomics, phyloseq, DADA2. Exciting work on data quality, ordination. https://t.co/dRsMaHuoQK
Elegant access to @ensembl transcript models in @Bioconductor by Johannes Rainer from https://t.co/E0mYsnc9T8 https://t.co/A3OWyr2Emg
Don Knuth at useR 2016 on literate programming. The origin of Sweave, knitr, vignettes… https://t.co/VQ6eV9xsTM
Don Knuth at useR 2016 on literate programming. The origin of Sweave, knitr, vignettes… https://t.co/nspZVJYf22
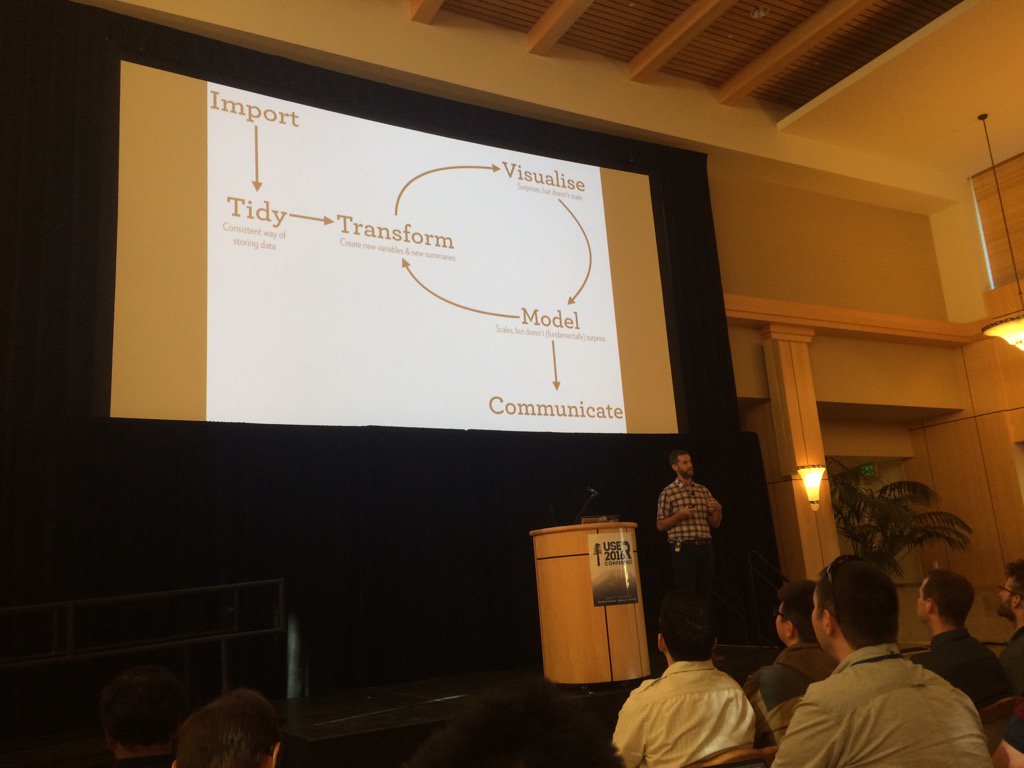
Hadley Wickham on data science. Only visualisation can surprise. https://t.co/gkDQmSqlF0
R at LHC/CERN - statistical analysis and optimisation of exabyte particle physics computing infrastructure 1/2 https://t.co/ZZDnpmNvaX
R is like a bicycle for the mind; Hadoop a containership. 2/2
Pertinent points by Deborah Nolan on modernizing statistics teaching https://t.co/sbQIj8jSAm
@rstudio conference Jan 13-14 in Orlando: https://t.co/Ld2fOwV7dX https://t.co/O6ikWC5vCg
Interesting point of view by AC Grayling appealing to the sovereignty of parliament over plebiscite https://t.co/XlH9vlrMyu
Postdoc Positions Evolutionary Single-Cell Genomics in Kaessmann-Huber-Arendt labs EMBL&ZMBH https://t.co/lVZQXGvQZD https://t.co/nBksxyIRcr
Assistant Director position at Centre for Molecular Medicine Norway (NCMM) https://t.co/F3NnfGfhUG https://t.co/v14TlXBvga
Getting ready for #csama2016 Statistical Data Analysis for Genome-Scale Biology https://t.co/EU8AbbhpM7 https://t.co/3kQRktfUre
@olgavitek Fantastic location, ideal for summer school: lots of chances to bump into each other and stunning nature. Great lineup you have!
#MyEuropeanMoment hiking in bilingual South Tyrol and getting lunch at a wonderful mountain hut https://t.co/QkPHbJ8wgd
Dinner table conversation: how do you distinguish a genuine scientific field from a citation cartel? https://t.co/yClZf3quoP
@areyesq … and now with corrected notation Spot the difference? https://t.co/O93zduSSzr
Bioconductor workflows are executable documents that explain end-to-end solutions in genome-scale data analysis https://t.co/D2UzC7v6ig
Open-source projects on github encourage small contributions from “outsiders”, e.g. documentation/typo fixes. 1/2 https://t.co/bzOUbLCI2D
As they gain confidence and standing, they can move “up” to become more involved. 2/2 https://t.co/9L0QOd03Jv
@mikelove Yes, “all Horizon 2020 projects have obligation to make any peer-reviewed publication open-access” https://t.co/9RtQYkvsgI
@chapmandu2 @BioMickWatson Phil – Junyan Lu said he’d do it. https://t.co/J7BhHhe10T
There is H-2020, for consortia, often “translational”, and ERC, for PI-driven basic science. https://t.co/weVOyumJiL
@hadleywickham 1. Assumptions are sufficient, usually not necessary. 2. Failure to reject does not prove assumptions are true.
ECCB Workshops now open for registration https://t.co/Sj0MhBDMJI – Clinical Bioinformatics as a Service https://t.co/u50H9w9pgw
@andreamrau besides the two transformations studied, would it be useful to include the obvious ones: log(n+c), VST, rlog, moderated CPM?
@andreamrau even with mean-centering (per gene, across samples)?
Stephen Hawking on our attitude towards wealth and cathedral projects https://t.co/BhNJOgY9Jm
@jtleek Freedom. Going to the bottom of things. The product (science) is valuable. PI job seems like running small business, many ups&downs
Germany is joining Elixir: Building international infrastructure in life science informatics https://t.co/1LkK23YxQV https://t.co/ts4fERM0iP
@NancyScience German government signed a contract required for joining. Next step will be the node application.
#firstsevenjobs painter newspaper boy, assembly line, locksmith, truck driver, programmer, researcher @SherlockpHolmes
@CFlensburg @genetics_blog Sufficient & necessary assumptions are not the same. Focus should not be assumptions but bottom-line performance.
@JennyBryan @hadleywickham @polesasunder sample(x) is the stringsAsFactors of permutating numeric vectors
@JennyBryan @hadleywickham @polesasunder S3 methods are the stringsAsFactors of namespaces.
@hadleywickham Subsetting named vector with factor doesn’t use factor levels, casts factor to integer… not sure that’s in scope of forcat?
@Bioconductor Developer Meeting 6.-7. December in Basel https://t.co/pysSblu8bp register now
My favorite unified interface to machine learning in R is
@groundwalkergmb @Bioconductor Lots of love -but this isn’t a biology specific activity, & Bioc has spent effort more wisely on other fronts
What to make of a bioinformatics postdoc applicant’s PhD thesis when all its figures are pixelized Excel barplot screenshots?#youcandobetter
fortunes clue Rcpp grid magrittr dplyr ggplot2 #7FavPackages #rstats
A call to value software as a research output (like papers). Software can be human-readable; analogy to math proofs. https://t.co/DITNzul7T1
@KasperDHansen See also CRAN package ‘xkcd’ for xkcd-style graphs incl. fonts https://t.co/5eYxFJqrRU
Interesting: large-scale survey of trends in usage of bioinformatics databases and software by literature mining https://t.co/nxgmO9xuTG
informal meeting of R users from the Basel area https://t.co/9ze8FNdctk
@timtriche @mikelove As Mike says, these are (squeezed and rotated) ECDFs - wouldn’t violins or beeswarms be better? https://t.co/iWCKpvMRmv
@mikelove @timtriche Point taken, but do we really want readers to parse sth so subtle out of a background of noise? I prefer more explicit.
@Malarky67 @timtriche @mikelove there’s a pressure to constantly innovate, invent. Result is often reinvention - at cost of clarity and ease
@ewanbirney But the issue is not the economy, it’s the many people who feel left behind. Nationalism is a time-honored ersatz mood lifter.
9000 R packages in CRAN: “We’re not just laying brick: We’re building a cathedral” A big thanks to CRAN maintainers https://t.co/XrGSNqe8Y7
Beware of visualizations that impose structure on the data rather than revealing it. https://t.co/M9RcyvqCvQ
Misconception. With good statistics you can do experiments that’d be impossible otherwise. Plus, you can do studies. https://t.co/xLFW95GF5d
A gem. Wonder what’s next … flat earth? Newtonian physics (i.e. not that EU-fangled relativity and quantum stuff)? https://t.co/CqohRVrA1W
My beach read: @HankGreelyLSJU’s The End of Sex- brilliantly presented account of the future of human reproduction.
@HankGreelyLSJU Nature goes to great lengths to protect the germline genome, we’ll need to do the same. Other cells than skin?
@hadleywickham Perhaps a problem for Empirical Bayes or AI? For <30 points, prefer geom_dotplot, _beeswarm: histogram is the wrong hammer
@genetics_blog # different defaults: > %in% function(x, table) match(x, table, nomatch = 0L) > 0L # default for nomatch is NA_integer_
From The Economist Espresso: Blue riders: the meaning of colour - at Beyeler near Basel https://t.co/SZuIsuACXc
Lesser men just dig a canal and let lots of boats sail around the mountain. https://t.co/0o2FaGMjUl
PI positions @embl https://t.co/jPWZi595f4 Great science, exciting international environment, lots of opportunities. https://t.co/itQQhAitSk
Perspective on the two pillars of truth in science publishing: peer review and reproducibility https://t.co/ixYuRMG08o by Thomas Südhof
One year as a data scientist at stack overflow by @drob https://t.co/CAcfjr79bJ Advice to graduate students: create public artifacts
In praise of introverts https://t.co/qrO4ziKJc1
Latest issue of R Journal https://t.co/R341hTgfFi computational geography, text analysis, gender prediction, mclust5 https://t.co/tQaYAQiwOR
No. It’s the basis of the scientific method. Theory needs to follow empirical observation, not the other way round. https://t.co/GwGHnFPApW
+1. Choice of RNA-Seq methods: base not on dogma or who shouts loudest, but on well-designed independent benchmarks. https://t.co/KkOt2juMRu
Audi or BMW? https://t.co/tYmMxJjvSY
Valid R: > ❤️ = factorial > 😀 = 8 > ❤️(😀) [1] 40320 https://t.co/z3NXXM3iKN
Good tradition since Plato and Hegel. https://t.co/CFWgPVUt12
Monument to the anonymous peer reviewer: https://t.co/fnDlmPj3VN #wonderfulsymbolism https://t.co/nceLk9huDb
Abstract deadline 15 Sep: EMBL PhD-students Symposium Life by Numbers - Towards Quantitative Biology 17-19 Nov https://t.co/DJbPVYV9lp
Best ask @DataProgrammers @lgatt0 & https://t.co/Q4JlcVQqo2 No follow-up date as of now at @EMBL, but would be great https://t.co/vHoAQHL3QO
But F is a good name for a matrix. There are few reserved words in R, type ?Reserved. #BeCarefulWithYourNamespaces. https://t.co/QS8cqxHTwo
@Bioconductor release schedule https://t.co/EmYdotAPGG Deadline for new packages for autumn 2016 release: 26 September!
“The Power of R” karaoke by Jonathan Crabtree https://t.co/nayyWzEbDS @ewanbirney @sjmgarnier https://t.co/ypcXZzW4JZ
@lpachter in both cases - mature technologies - multiple criteria for what’s good - ready for disruption (long reads, electric, self-drive)
Codesharing in science:we’re leaving world of narratives that depend on computation w/o supporting digital artefacts https://t.co/mzWoLLpg78
Using FDR in your analyses? Don’t loose power by treating all hypotheses the same- smarten up with domain knowledge https://t.co/aczz02p1tZ
French trains zipping at 320 km/h through the sunny countryside are amazing.
Phd defense by the brilliant Elsa Bernard at Institut Curie in Paris Author of flipflop https://t.co/rwXtZQPd5U Congratulations! https://t.co/6tcEzK4bqH
Don’t be a money pump (by buying phone insurance, extended warranties, collision damage waiver). Tim Harford is “The Undercover Economist” https://t.co/yEdfQgwCQw
Rising income inequality in science pitches insiders vs newcomers, old vs young https://t.co/3tvuh4dtEd
Can you spot the Epimenides paradox: https://t.co/pUUkBOCt4k
tufte and tint - two new R packages for beautiful @EdwardTufte-inspired Rmarkdown layout https://t.co/807eGA01D7 https://t.co/HmhG9vQ7Ow
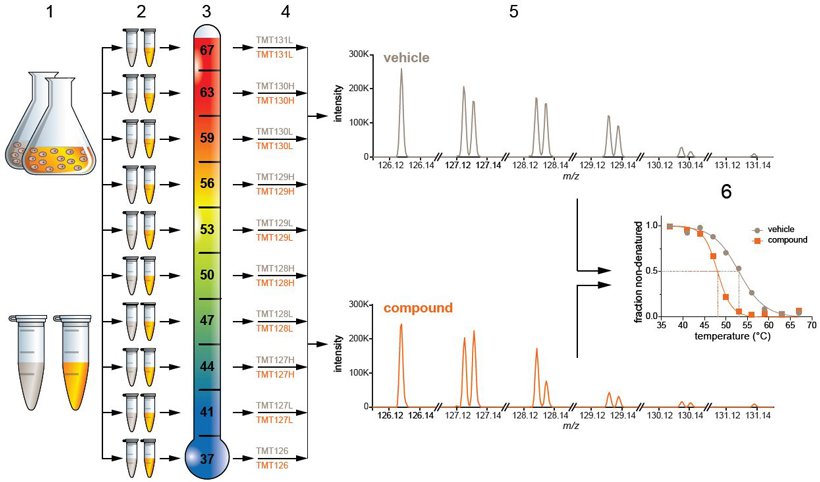
Off-target activity of HDAC inh panobinostat - discovered with cool new mass spec based high-throughput assay, 2D thermal proteome profiling https://t.co/18WfTa5gUI
Post by @jtleek on emotions and tempers in arguments between data producing scientists and statisticians https://t.co/NBTMPmLvy5
Economist article on “ethic of conviction” vs “ethic of responsibility” https://t.co/OJWIDYBpYz
@LorenzAdlung It hadn’t occurred to me that this wasn’t obvious.
@ewanbirney Sad but true. Seems frivolous to paralyze ‘the West’ about such stuff in times when the world has several real problems.
At 21, I did Interrail tour of UK&Ireland. At 22, came to study in Edinburgh. Easily one of my best investments ever https://t.co/I5EyLsGMtO
@ewanbirney @dermitzakis Probably well-intended but definitely fallacious data normalization.
Sites like CRAN or Bioconductor make it easy for scientists to distribute their software through package manager. Daily builds, unit tests. https://t.co/hGvvEUa1D3
@bjoerngruening @BioMickWatson @genetics_blog No “would”: previous releases till back to 2005 are archived&available https://t.co/glyhkOEKUn
@bjoerngruening @BioMickWatson @genetics_blog I just posted a link for you with the previous releases. Can you elaborate your problem please
@bjoerngruening @BioMickWatson @genetics_blog They’re here: Category 2.36.0 https://t.co/i6GnNGEgeK DiffBind 1.16.3 https://t.co/emOcTa21uk
@bjoerngruening @BioMickWatson @genetics_blog Agree system far from perfect, efforts like bioarchive are great. Also https://t.co/QE8bspjsoB
@bjoerngruening @BioMickWatson @genetics_blog Why use non-release version? If desperately needed, rebuild from svn https://t.co/VOFvl4miem
Great interview with J.J. Allaire, founder of RStudio and modernizer of R https://t.co/dA3ra96Cpw OT @hspter
Great speaker lineup at #denbi Symposium Bioinformatics for Human Health and Disease 7-9 Nov in Heidelberg https://t.co/FxqMi1QpKb https://t.co/QfIPK3XTZh
Statistical Challenges in Single-Cell Biology - Workshop in Ascona/Monte Verità. Submit your abstract now: https://t.co/5MNIjMKqKH
These will be exciting data. I wonder whether a cross-species, evolutionary angle would make them even more useful. It did for the genome. https://t.co/5e7g8MxyZT
#Rstats trap #1729: x[ -which(x < th) ] to select all finite values >=th.
Except if no element < th, in which case you end up with nothing
which removes the NAs: > 1[NA] [1] NA > 1[which(NA)] numeric(0) https://t.co/37p30sC0OJ
We need more data-driven journalism, for many areas of life, not only polls. Report objective data, not anecdotes. Good career direction! https://t.co/9hHufUOrqw
@StephenEglen @lgatt0 @MMaechler apply is just wrapper around for-loop;modify yourself? Not easy to generalize to distr.mem.,consider rhdf5?
I wish the term “adjusted p-value” never existed. FDR applies to a set of hypotheses. It’s not meaningful for individual hypotheses.
@dbkell BH is great, but assumes hypoth. are stat. exchangeable, which can be inefficient. Modelling non-exchangebt is harder but worthwhile
@mikelove @notSoJunkDNA Nomenclature problems are the hardest ones in science. “q-value” is fine.
@MMaechler @hadleywickham I agree. Still keep seeing it in code I review, hence a reminder.(It’s superficially smart b/c ability to drop NA)
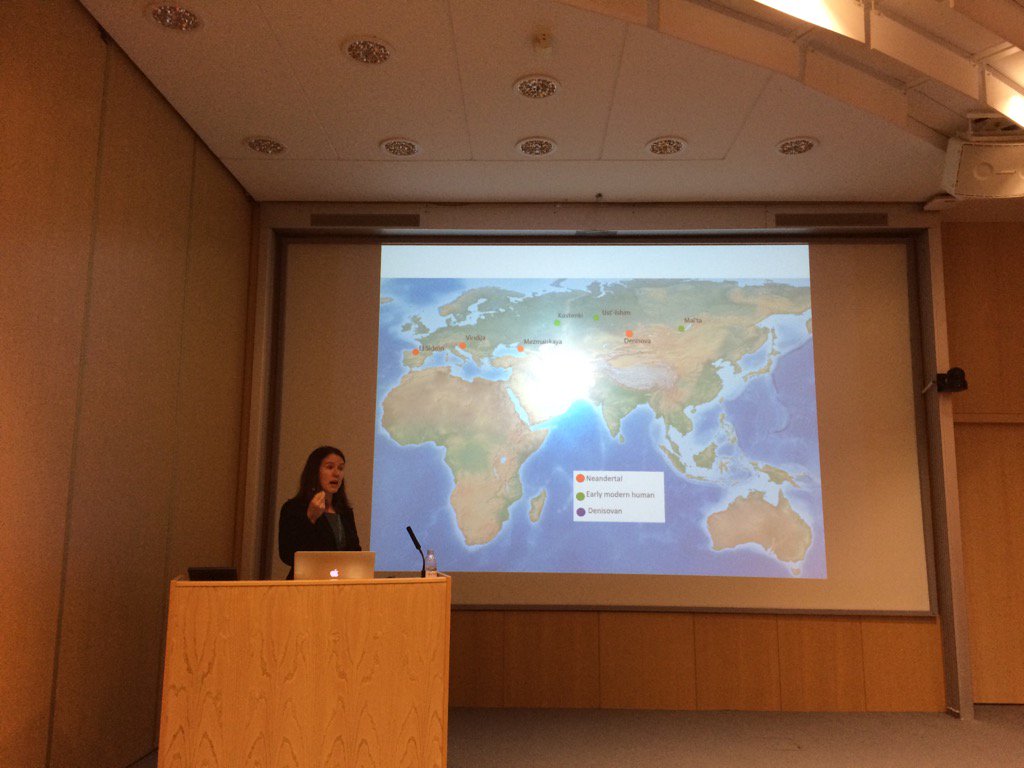
Janet Kelso talking at EMBL on genetic admixture between modern humans and Neanderthals https://t.co/pAzL5oLodq
How not to analyse data. The data don’t fit a line, but line is fitted anyway. Slope depends on arbitrary choices of data processing. https://t.co/lhNm7oRSYS
@johnstorey So it has lost control over you.
DESeq2 vignette now easier to use online in HTML (Rmarkdown) instead of PDF: https://t.co/h1LM4J6xxF Thanks to @mikelove
Removes imports and exports from all installed package NAMESPACEs. Replaces all mathematical algorithms with precomputed wrong solutions. https://t.co/FLEq2BWhx4
Amazing if true. Ctrl+Z: Silicon Valley leaders U-turn on Donald Trump https://t.co/GDymraNani
In these times we need good, independent media. Support them by paying a subscription, don’t only rely on “free” online offers.
Normalization is important for ’omics data. But doesn’t replace quality control. Should not normalize bad, outlier data.
Prof in labcoat on mainstream TV news converting tumor GEP data into music. Not sure whether to wince or be happy about science in public.
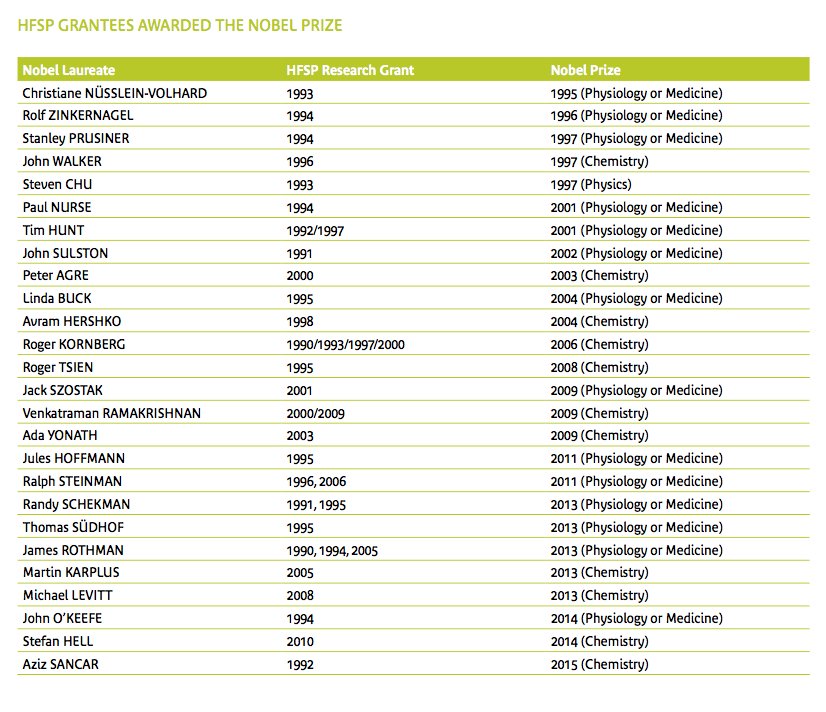
Cool & hyper-successful: @HFSP international research funding, postdoc & young-PI fellowships, project grants https://t.co/DMA2InKK0e https://t.co/2aSFNLtxHX
@neilhall_uk Except that the ‘Independent’ article is another example for sensationalist fake news. Needs push back.
@DrAnneCarpenter Not directly what you’re asking for, but related: Transparency International https://t.co/9nj7IhVv5K
Bioc Workflow: Genomic variant annotation workflow for clinical applications. Thurnherr et al., @F1000Research, https://t.co/wNiKzih0ng
Prime minister of Baden-Wuerttemberg state at @embl https://t.co/LxTkgn8rMw … https://t.co/Bvwn6ZZ2iC
Exciting program for Bioconductor Developer Meeting Europe #EuroBioC2016 in Basel next week https://t.co/pysSblu8bp
@ewanbirney @pedrobeltrao Until biologists stop outsourcing a key task -ranking papers- to a small gatekeeper club of elite journal editors
@ewanbirney Yes of course. Could’ve said “scientists”. @segregatedxx: what is the point?
Welcome Slovakia and Hungary to @EMBL! #scienceisglobal
A database of single-cell RNA-Seq datasets, extremely valuable for method development & benchmarking. And for re-analysis, biol. discovery. https://t.co/L0WI7xU3OV
Curated Metagenomic Data of the Human Microbiome - new @Bioconductor pkg by Levi Waldron et al. https://t.co/N2o0v3ICIX
@ewanbirney Is there already a date and place for #EWGtarget 2017?
Benchmarks are most useful if by disinterested 3d party. But by competitor also interesting: they try to beat your method &find weaknesses
@wolfgangkhuber Just please don’t use something computed by your own method as ‘ground truth’.
@chapmandu2 @ewanbirney Yes. #EuroBioc 2017 will be in Cambridge 4-5 December
@kara_woo @STAT545 We need a package to stenographically embed (& extract) associated #rstats code in jpegs, pdfs
@ewanbirney Using base-R data.frame? Consider data.table https://t.co/0Y0btEymnM Also tibble can be more mem-effc’t https://t.co/0QDQMQJYx6
Airmass inversion - thick fog in Rhine and Neckar valleys, beautiful sunshine on @embl hills. https://t.co/lug5GuI8xh
PhD-level staff position in computational modelling and quantitative biology @embl - exciting career opportunity https://t.co/mtGj86VR76
When your readers contact you about downloading a massive dataset directly from you b/c the public repository is too clunky #mustdobetter
Decoupling of TADs & chromatin compartments: Two indep. modes of chromosome organization revealed by cohesin removal https://t.co/kkeQ5Z2JbC
@mikelove Why not depend on it?
Swiss scientists re-join Horizon 2020 programme https://t.co/9VD8qBRbp7 https://t.co/qw4iKUE5tO
@AliciaOshlack -False precision fallacy https://t.co/4TUcqJ1xYI -The analysis is likely worthless as it started from unrealistic 0-hypoth’s
Who could ever be against that. https://t.co/Kgu3EuWmUO
@lpachter Guess that’s irony? Size of visible universe in electron radii: O(10^42). Silly to peddle such probabilities @AliciaOshlack
@ewanbirney @lpachter @AliciaOshlack But that is very unhelpful. Effect size and p-value are (in general) two orthogonal things.
@notSoJunkDNA @ewanbirney @lpachter @AliciaOshlack Best report something else, from more interesting analysis. If have to, use upper limit.
@greg_slodkowicz @lpachter B/c aim is to make statements about the real world, not (only) about mathematical models of it.
@greg_slodkowicz @lpachter E.g. if your dice has a small imbalance and its probabilities are off by 1%, that’s x2.7 after a 100 trials
@ewanbirney @lpachter @AliciaOshlack Nobody doubts worth of p-values! Just ridiculously small ones that exceed model uncertainty / assmptns
@greg_slodkowicz @lpachter Of course. But “very low” is not the same as a specific ridiculous number. https://t.co/4TUcqJ1xYI
@greg_slodkowicz @lpachter They could. And if that were the main result they’d report from these data, they’d be poor analysts.
@lpachter Oh come on, stop trolling. They did a lot more with these data. And yes, that false precision in GO enrichment I’d avoid next time
Many good points. When you only have a hammer (testing), everything looks like a nail. Good to employ a wider set of tools. >> https://t.co/4LlMF3VuEG
But not throw out baby w bathwater. Mult.testing as intermediate screening followd by separate validation is diff’t from single “final” test
@wolfgangkhuber And, as @SherlockpHolmes points out, key is transparent reporting of raw data and performed analysis (e.g. Rmarkdown script.
@lpachter We all agree that p-values are & GO analysis can be useful. My point is not to report single ridiculous numbers but an interval.
Not impressed by latest Apple software. iOS10.2 made old iPhone5s unusable. Brand new MacOS10.12 laptop keeps crashing apps & iTunes a pain.
On tSNE: https://t.co/5ypUYUE752 I like the uniform distribution in unit-cube, or set of all equidistant points. Beware of parameter choice.
@ventutech It used to be that everything “just worked”. Now drivers (video, sound..) and apps are as buggy as in any other OS #expensivejunk
Advanced R Programming Course in Heidelberg at @EMBL 3-4 April with @lgatt0 & Robert Stojnic https://t.co/eIRJTEagor
Statistical Challenges in Single-Cell Biology, Ascona April 30 to May 5, 2017 https://t.co/5MNIjMKqKH … Great speakers, location https://t.co/7S97MYCJVb
@ewanbirney It’s not a null-sum game. Everyone looses. It’s also not really about economics Sad context: authoritarianism growing everywhere
Who knows a good viewer for microscopy images in HTML5/JavaScript (embeddable)? Scrollbar for z-stack, time, mult.color channels wd be nice
@Fjukstad Great, let me -and twitterverse- know when!
@notjustmoore @bioformats Want something that runs purely in browser, just HTML5, client-side JavaScript Image format: whatever, can convert
New paper: “Covariate-powered weighted multiple testing with false discovery rate control” The theory of IHW https://t.co/0c7Zh7zvEV https://t.co/oerN4751bT
5 travel bursaries for Ascona Workshop Statistical Challenges Single-Cell Biology https://t.co/5MNIjMKqKH thks SOUND https://t.co/fBjBVXYe6n
Ironic how this is now creeping up in two leading countries of the West. Important not to “tune out” about what’s truth, or what’s knowable. https://t.co/4dfXyJe74s
MASAMB 2017 April 3-4 in Vienna: Mathemat & Statistical Aspects of Molecular Biology. Great conference esp for young researchers & newcomers
Cool postdoc at FMI in Basel in live cell imaging, HCS, quantitative imaging, mechanisms of organoid formation https://t.co/AHKyFo0up1
Glamour journal asks for “quick” 3rd review if a fully computational paper is rigorous, after no computational reviewer was asked before
This is great: science becoming a profession that allows people to move in and out, rather than a vocation with expectation of linear CV. https://t.co/MIdH1mnbkV
tidytext (#Rstats) sentiment analysis of Trump’s tweeting: more angry/sad/negative now than any time in past year https://t.co/tiIY45Gk9t
This is excellent https://t.co/JfGoEOKdoq
Advanced R Course 3-4 April at EMBL Heidelberg Instructors: Laurent Gatto, Robert Stojnic (Cambridge) https://t.co/VvERQ0HeQ8 5 places left
Faculty Position (Group Leader) at @EMBL: Genomics Technology Development https://t.co/UtETxbqOJm https://t.co/AlzxRjNhGF
@ctitusbrown @lgatt0 Bringing self-citation to an entirely new level. Also useful for creating cycles in the citation graph.
Registration now open for Statistical Data Analysis for Genome Scale Biology Brixen SouthTyrol 11-16June https://t.co/gZjBTD8Ndc #csama2017
Rhineland carnival floats were having fun today https://t.co/2K3t6Z3wBv
@mikelove Theory, concepts, abstractions last longer and are better use of quality time. Instill respect for wrangling, give pointers, labs.
Excellent career opportunity for first PI position.
Great resources and outstanding environment. https://t.co/Gqk2uNQ0tj
@mbeisen Some is gratuitous, but valuable to be able to re-run w/diff. parameters, resample, provide to 3rd parties as reproducible workflow
Wow…. 85 vs 2 vs 5 pages https://t.co/uamMlnKlAY
@mikelove Roadblock is a good metaphor: it’s also good to know where you want to go and what the road network is.
Talked to Iranian-born scientist working in US who has to cancel speaker invitation from Europe for fear of no return. Disgusted.
@ewanbirney Visualizations ought to show structure that is in the data; not make up structure.
Advanced R Course 3-4 April EMBL Heidelberg Instructors: Laurent Gatto, Robert Stojnic (Cambridge) https://t.co/VvERQ0HeQ8 3 places left
Overcoming the limitations & confusions from non-standard evaluation in dplyr: https://t.co/0eXtg0EGeg
Friday afternoon quiz: What does this R code do: library(“magrittr”) %<>% %<>% %<>%(%<>%)
@hywelowen @david_colquhoun Perhaps she’ll feel better with c^4/G ? (GeV/c^2 is a unit of mass, not weight.)
Advanced R course at @EMBL by @lgatt0 and @rbstojnic https://t.co/4wMaHEXkyX
ggplot graphics with emojis using the emojifont package https://t.co/rb1aeR7uTE https://t.co/k7xX9WNjgO
@Bionductor conference 2017: https://t.co/WXsUKENiBT - including a call to contribute workflows through F1000Research / Bioconductor channel
de.nbier https://t.co/1pdRoqRxhi thanks to @MalvikaSharan https://t.co/beo4Hqi7I5
@AedinCulhane @eLife @Bioconductor @rstudio Wow - this sounded like science fiction but is now for real: submitting manuscripts to the journal in Rmarkdown - @eLife
@lgatt0 @AedinCulhane @eLife @Bioconductor @rstudio @F1000Research @github https://t.co/UY61kwzj23 functions to ease the transition between Rmarkdown and the LaTeX used by F1000R for Bioc workflows, by Mike Smith
@OliverStegle @AedinCulhane @eLife @Bioconductor @rstudio Maybe - though some of the more arbitrary journal format choices seem a thing of the past, compared to, say, author style or article type.
@OliverStegle @AedinCulhane @eLife @Bioconductor @rstudio Check out @andrzejkoles’ BiocStyle https://t.co/If2UTpdngX that provides one good choice for such a template: e.g. https://t.co/YG8E7YQho4
@AedinCulhane @OliverStegle @eLife @Bioconductor @rstudio @andrzejkoles BiocStyle works with bibtex / .bib files, so I presume the answer to your question is “yes”?
Great place to work: https://t.co/2gaZau7W6E
I’m always amazed how many people are not aware of the difference between sufficient and necessary assumptions.
March for Science Heidelberg https://t.co/Viz13TS0gC
@ewanbirney And that you can indulge thinking about 0s in ENSG-IDs and not need worry about a rumoured scaling issues of R with these ‘big’ data
@ewanbirney It’s a recurrent meme that R is slow (https://t.co/hRd3Q2CtOb) but as @mt_morgan shows the data are conveniently handled in R/@Bioconductor
What’s wrong with UK media? Top trending topic for two days on https://t.co/nr0nzzm6n0 is the fascist who came 2nd https://t.co/xwoJA9eLKe
@Bioconductor 3.5 is released
88 new software packages, 1383 altogether 315 data, 911 annotation packages https://t.co/jSLOcAQyBk
On the way to Ascona Single Cell Workshop, train racing across the Alps through the amazing new 50km long Gotthardt tunnel https://t.co/Zgj8E0uIqc
Versatile toolset for functional data analysis, e.g. time courses, drug response, melting curves, … https://t.co/S9ttpkWGh4
Great biography of this amazing scientist, a 19th century Indiana Jones: https://t.co/h0MEI4pi5U https://t.co/luGcl5mO9f
Reviewer for paper w @areyesq: “they used non-standard analysis method” means: “they used method that I didn’t invent” #BrokenPeerReview
Excellent bio-pic of Marie Skłodowska-Curie.
F-rated.
https://t.co/unKkTlFIFn https://t.co/sqSNC3zy9X
Personnes rationnelles de France, votez! L’isolationnisme est se retourner, il faut aller de l’avant.
Je vais avoir une verre de champagne sur ça.
Translating genomics and bioinformatics research into healthcare tools and services https://t.co/ILu2uImj1Q https://t.co/2YD1TpPC1m
@ewanbirney What do you think non-UK Europeans can do concretely? (assuming that view of science being international is uncontroversial to their govts)
@ewanbirney This is so uncontroversial, and evidence is not the bottleneck. That’ll be the increased hassle and obstacles after loss of single market…
@mikelove on using RNA-seq DE methods to detect allele-specific expression https://t.co/nErOpNiZ0A https://t.co/spYE6jUsZF
@torstenseemann … and the limitations of experiment techniques/ measurements
Unpleasant experience with @bookingcom: at time of booking they claim ‘free cancellation’, once booked, cancellation fee is the full price
A committee is a group that keeps minutes and loses hours. #fortuneoftheday
@iacus @lgatt0 And another view from near Brixen, the site of #CSAMA 2017 https://t.co/M8qHXxpZ7L
Junior Career Fellowship for MD students to do science projects - Heidelberg Research Center for Molecular Medicine https://t.co/UEcG6uS068
Coffee break at #csama wit @mt_morgan @jotsetung @lgatt0 Vince Carey https://t.co/hwbFif32iG
Idea: Rexit - reimplement all of CRAN and Bioconductor in BASIC, in 2 years.
@cwcyau @mikelove ..or stuff we don’t want to model “Other” is good, “Fluctuations” a physics-inspired alternative 0-centred not needed/ambiguous if nonlinear
From Cologne: cyclist parks bike on car lane with the kind of excuse drivers use to park on bike lanes https://t.co/NYyx9JfEde
Brooke Morriswood reviews biological journals like restaurants: https://t.co/j5SDSfVDf8 (via Angela Andersen of Life Science Editors) https://t.co/4H0HTez4V2
@AliciaOshlack @JovMaksimovic A case for a polite description of the experience at Pubmed Commons? https://t.co/L4K662sLC8
1/2: “If Boyle et al. are correct, then hypothesis testing is not an appropriate statistical approach for GWAS.” https://t.co/Ets2Log3B7
2/2: So multivariate regression and classification? Btw, hypothesis testing with FDR may still be useful for feature selection.
Posts in @PubMedCommons appear right under the abstract of the paper after a Pubmed search. Authors can then respond, too.
Question: do women get the comment “Nice talk, beautiful work” more often than men after a conference talk? Is it condescending?
@tslumley What’s a use case for that, i.e. when isn’t duck typing enough?
@robinson_es @chendaniely See also output: BiocStyle::html_document2 with lots of features for writing fully fledged academic papers https://t.co/tnxEuhCAM9
If 0 is just another number in the NB distribution, does it really need a special name “dropout”? Seems distracting & unfair to the 1s, 2s.. https://t.co/lGY8Ps5wA4
Right in time for useR! 2017 in Brussels: new issue of R journal https://t.co/VSjUAF26hN
@neilhall_uk Making education expensive is a time-tested way to limit social mobility … both ways.
Redefining the kilogram - using universal constants instead a piece of metal in a city on Earth https://t.co/sdRx5HHuqQ
Erasmus -the international student exchange program- is just great in so many ways. https://t.co/qyalAtGEus
Same argument for EMBL-EBI Cambridge vs Hinxton…? https://t.co/SJG2sU9RV6
Do you mean strand-specific or sequence-context-specific (what @imartincorena refers to)? https://t.co/W2K3LLLU0i
Opportunities to start your research group at @EMBL - fostering diversity, work-life balance and excellence https://t.co/5Hv0ogtnp4
@imartincorena Sure, but can’t there also be strand-specific errors not caused by sequence context (but instead, e.g. by software or data management bugs)?
AaronLun at #bioc2017: analyzing the 10XGenomics 1.3 million cells data in R on 8GB Laptop using scran and beachmat https://t.co/674XztSY5W
@jcbarret @jcbarret For correlated phenotypes, could do dimension reduction first? Fewer tests -> less stringent Bonferroni (1/2)
@jcbarret With FDR, important to realize it’s an average property of a set of rejections. Local fdr of indiv. tests can be higher, lower 2/2 https://t.co/u5TBKOBGQD
@jcbarret So there are many ways to achieve same FDR with different sets of rejections. This is what underlies co-variate informed FDR-methods, eg IHW
@jcbarret @psychgenomics Ordinary Benjamini-Hochberg works well when there is dependence.Hardly worth worrying about.Would break down only for pathologic situations.
Apt analysis of the state of biomedical science (somewhat ironic choice of publication location) https://t.co/InLxdICOcQ
Elegant interface to #rstats arrays that are much larger than memory in HDF5Array and DelayedArray https://t.co/Leac154tal by Hervé Pages
@jtleek Great data collection! -log-scale? -use density estimator that does not go <0? -what is to learn from stratification into fields?
Rahul Satija on finding common low-dimensional explanatory manifolds at #bioc2017 https://t.co/xFxvNswVAG
Excellent talk by @elhamazizi on BISCUIT - simultaneous normalization & clustering applied to breast cancer single cell data #bioc2017 https://t.co/p5jCkLZWXc
John Storey on using informative covariates for optimal ranking of tests in multiple testing. #JSM2017 https://t.co/z5uyLEM7qS
Consider application of Goodhart’s law to glam journal publishing: “When a measure becomes a target, it ceases to be a good measure.”
My slides #JSM2017 session “Learning from External Covariates in High-Dimensional Genomic DataAnalysis” 8:30 CC-329 https://t.co/NpwOTqi0rc
Britta Velten at #JSM2017 on co-data informed penalization in high-dim regression. https://t.co/6Rrdgga2mQ
.@groundwalkergmb New built-in vector representations in R give huge performance boost: sequence, RLE #JSM2017 https://t.co/HqYGCGo15H
Great to readjust timezones by mountainbiking in the wooded hills around Heidelberg after great trips to #Bioc2017 #JSM2017 https://t.co/KzMP3ySar9
… and Bioconductor Europe conference Cambridge UK 5-6 Dec 2017 #eurobioc https://t.co/tBMKoRqXaA
Cicero trilogy by @Robert___Harris - vivid account of the Roman republic and the triumvirates. My #BeachReading
Now “A Crack in Creation” by Jennifer Doudna https://t.co/dhiUmdZEm0 https://t.co/mEm5iEypX6
Use the new BiocStyle to create HTML vignettes that look good & can have all the layout features of academic papers https://t.co/uYpht4j2SQ
EBImage package now offers the option to view images in vignettes in a Javascript-based browser https://t.co/mtmnqLesUD
@StephenEglen Yes, see the example mentioned by @andrzejkoles - https://t.co/4aAbBLgmTy
@StephenEglen @andrzejkoles See https://t.co/5Vy6IynueB - just choose between output: BiocStyle::html_document2 and pdf_document2
@HankGreelyLSJU History never exactly repeats itself, yet: real problem wasn’t Hitler, but efficient people under him who he empowered with his popularity.
Bavaria to spend 10^9 Euro on a new university, in Nuremberg, 100 new professorships https://t.co/TmIHY0Y5H0
There is no place for bullying and online trolling in bioinformatics. https://t.co/9CYSGjmR5G
A familiar pattern It’s convenient for those who set the deadlines to come back to work on assessing submitted grants after /their/ holidays https://t.co/oVnaYuC0hf
All @Bioconductor packages now have a DOI. E.g. https://t.co/rPVyKjeWcy Thanks @seandavis12 !
European Bioconductor meeting 2017, 4/5-6 Dec, Cambridge UK For all who use or contribute to Bioconductor or want to https://t.co/5ateL2BVcM
There will be a new high-resolution microscopy centre at @embl Heidelberg. - Thanks to federal and state funding signed today. https://t.co/wg0IchmTqC
Bioinf PI positions at EMBL-EBI in Cambridge. https://t.co/7m9IwhBS3v EMBL hires on potential, not experience. Great diversity workplace. https://t.co/xn4FEZVaGi
@MelanieIStefan @StephenEglen There’s a lot of support to increase diversity. But we’re not yet where we want to be. This is work in progress.
Third Asia-Pacific @Bioconductor Meeting SAHMRI, Adelaide, Australia Friday 17th November 2017 https://t.co/DVjJNqaZEH
Full Professor Computational Genomics / Director Inst of Bioinformatics & Systems Biology Helmholtz Centre Munich
https://t.co/ElP9r7pCEn https://t.co/vcF1tVtBpL
Publish reproducible manuscripts as peer-reviewed articles - announcement by eLife: https://t.co/oSnD80JLOu
Professorship in Multi-Scale Modeling / Systems Biology at USC https://t.co/rostfG2G6V
Professor / Asst Prof (tenure track) positions in Vienna, Austria at multidisciplinary IST https://t.co/rpWIJD9StF
Statistical Methods for Post Genomic Data Annual workshop,this time beautiful South France Montpellier 11-12Jan2018 https://t.co/M3r2cBWzet
Looking forward to when the font sizes in all panels in all figures of a paper draft are the same.
@MagnusRattray Me too.
Workshop: Post-selection inference and multiple testing 7-9 Feb 2018 in Toulouse https://t.co/GpvGZI92n5
vsn paper proposes f(x)=asinh(a+bx), with a and b estimated from data based on additive-multiplicative error model https://t.co/IyqN4MuGHh https://t.co/12TurO3MQO
@arjunrajlab …and DESeq paper does analogous for Gamma-Poisson (neg.binomial) error model: https://t.co/VbqwKKBKM7 https://t.co/RRGq3m98e5
@arjunrajlab Btw, the variance-stabilizing transformation for Gamma-Poisson data is not asinh, but a function that interpolates between sqrt and log.
Quite a well-written CV: https://t.co/NXOcWrn3qz
Reminder that if we want quality news reporting, we need to fund it.
It doesn’t take much. Many have reasonable subs fees. Personally I support NYT, WP, Guardian, Economist, Spiegel, SZ.de, Welt, RheinNeckarZt
removeBatchEffects seems a terrible name for an analysis if the batch effect is that the assay didn’t work well for some of the samples.
@BioMickWatson Many biologists are excellent coders, and vice versa. All programmers use GUIs sometimes.
Ouch, I realize this could be read in two ways, I only meant one of them:
Being a good biologist is unrelated to whether or not can code https://t.co/pHqoc6gd33
Clarification: there’s lots of excellent science w/o computers. How to read this: being a biologist does not imply not being able to code.
@benoitbruneau The statement was something positive. It could also be read the wrong way. This, and any offense taken, was not intended, sorry for that.
Start of Stanford-EMBL Personalized Health Conference #ph17 https://t.co/aAU5Lfdxp1
Dorothee Nickles: predicting response to PD-L1 blockade from molecular profiles https://t.co/HTIWQuwZco
@ewanbirney @cdbustamante Isn’t that the point of insurance?
Britta Velten on Multi-Omics Factor Analysis - an extension of sparse PCA to multiple matrices https://t.co/KqxKyRZd20
Great collaboration with @OliverStegle and Ricard Argelaguet
@cdbustamante @ewanbirney Good points. Also, insurance model is adequate for sick care, subscription based model might be better for health care.
150th birthday! If you’re interested in some of her life, here is a passionate movie: https://t.co/0Cofe8C3b5 https://t.co/WVfzQZGuLb
Multi-Omics factor analysis – and application to a large study of blood cancers https://t.co/aq4RSyQWuD https://t.co/QBjRc53vRi
@embl conference Single- to Multiomics / Data Integration this coming week Sunday-Tuesday: https://t.co/720emUUtHT @zauggj
When there are multiple dimensions of author contribution and embedding into 1D, ordered list, is mathematically impossible (and btw, pointless). https://t.co/Yacf4CrGUi
Daniela Simancas & Christian Mertes present PatientBoard tool: helps physicians elucidate rare metabolic diseases using multi-omics, at SOUND meeting https://t.co/78gMXcd0UX Next step: use @Bioconductor Soundboard architecture. https://t.co/8Bdp2iv5bx
Alex Bertram @bedatadriven presents impressive progress with Renjin, a JVM-based interpreter for #rstats https://t.co/dQMJnYngXu
Impressive…. From: “Using Renjin as an R Package” (https://t.co/PreAyK0nzx) https://t.co/524F6mYic2
@_ms03 Vectorization is surely the best whenever it works - but sometimes the explicit iteration is easier. This was a toy example - what’s your suggestion for vectorizing it?
- and (2) are true, and perhaps especially for production; but (3) it’s good to have options, and (4) it’s fun to explore new avenues. https://t.co/QDIW4b9OTN
Flight of the Starlings - beautiful self-organizing dynamics of autonomous agents https://t.co/malbkx1ngC
@klmr The R interpreter is fine. Idiomatic R would use vectorization. There has been much progress recently, incl. ALTREP (https://t.co/HqYGCGo15H) & dplyr/etc. Still, many more optimizations are possible, as @bedatadriven ’s work shows, not only time, but also memory footprint.
sum(as.numeric(seq_len(1e9))) needs to allocate two long vectors, 12 GB altogether. https://t.co/6vk6FkTcNC
@bedatadriven @bedatadriven ’s slides: https://t.co/AgHptOE74R - Renjin update - Optimizing a HDF-intensive workflow
Bioconductor Europe Meeting 4 Dec 2017 in Cambridge: SIG Bioconductor for large-scale single-cell ’omics data analysis https://t.co/I28QqKPgjI
R/Bioconductor packages for single-cell genomics data analysis…: https://t.co/zko0NCBGZe
Jovial talk by Jacques Dubochet on the early days of cryo-EM of biomolecules in water at EMBL https://t.co/sIGHlN5Pie
Memorial Symposium in honor of Bernd Fischer Computational Genome Biology Friday, 1 December, 13:00 – 19:00, Heidelberg/DKFZ https://t.co/qzKS5X57ze
@davisjmcc Not encouraging at all. I’m puzzled why scientific community is OK outsourcing so much power (careers, grants, research agendas) to small club of ‘glamour rulers’ with secret, intransparent decision processes (..and tolerance for random acts of cruelty)
@davisjmcc Undoubtedly useful to learn to live with the current (publication) system. Encouraging: to dream of a better one.
Re impostor syndrome, good point. Good to read biographies of great and famous across history - look beyond twitter and current media hypes…
Way ahead of his time: Isao Tomita, pioneer of electronic music. Pictures at an exhibition (1975): https://t.co/Ha3gK0CCK7
Equal rights between women and men - the grand cause of his 5-year term https://t.co/0n5XCPyui0
It is never too early to start writing up that paper manuscript or thesis. Only once you write, you realize what you should have done.
@markowetzlab @PaulFlicek Real biologists have all sorts of talents, and it absolutely does not matter whether or not they program. That sorry tweet you’re referring to was a poorly worded response to people implying that all biologists are not able to program.
A great collaboration with @areyesq: Alternative start and termination sites of transcription, not splicing, drive most transcript isoform differences across human tissues https://t.co/PjSflG4Gc9
Well said: statistics is not a cookbook of recipes. It’s about asking the right questions, and figuring out how to make good decisions. https://t.co/jOaH4ogiYY
28th annual MASAMB workshop: Mathematical and Statistical Aspects of Molecular Biology - bioinformatics and statistical genetics Univ. St Andrews, 19/20 March 2018 https://t.co/yEPTw33ZKo https://t.co/qFQf7fhg3d
Replytoreviewers_final_reallyfinal_v09_sg_wh.docx
Sophie Rabe at #EuroBioc2017 on high-throughput cell phenotyping of leukemia-stroma co-cultures and drug screening https://t.co/86ORr8kx4V
EBImage https://t.co/0HSUZ10glC maintained by @andrzejkoles - many recent improvements in documentation and performance https://t.co/GbMJQAYCP8
Lukas Weber on clustering cytoF data and @gosianowicka’s workflow https://t.co/MTkNQ8lkqn - work from @markrobinsonca’s group in Zurich https://t.co/N2Wr0Uymcf
karyoploteR: versatile and beautiful plots of data along the genome https://t.co/Ird6wv8Lzr #EuroBioc2017
This looks like an immensely useful resource for anyone working in signaling: wyciwyg://4/https://t.co/J9HdP9cmrl
Drug-perturbation-based stratification of blood cancer by testing ex-vivo responses to a panel of cancer drugs, and multi-omics characterization https://t.co/LFUOSqC36d
Thanks to all coauthors for a monumental 5-year effort with many ups and downs, many nights and weekends…
Great! The data & Rmarkdown scripts that reproduce all analyses in the paper are here: https://t.co/LU90uAiGTz Hopefully soon also as data package in Bioconductor @andrzejkoles https://t.co/NTCPMvWeMM
New Independent Research Group positions in bioinformatics / comp. biology at Institut Pasteur in Paris https://t.co/5i6RFtRtDt
New Position at @embl: Project Manager - Heidelberg Center for Human Bioinformatics https://t.co/VYhjfeYhG0
That’s a cool postdoc project/position https://t.co/EQtKQyotXt
Today we are celebrating the birthday of a social revolutionary Jew born to a mother out of wedlock and brought up in a refugee family fleeing for his life. From @bjoerngrau
#itunes has such a useless (buggy) user interface. I do not recommend wasting money on renting or buying content with it.
Outstanding list of instructors at the Course in computation and statistics for mass spectrometry and proteomics May Institute: 30 April – 11 May 2018, Boston https://t.co/Pg90Ote4XH by @olgavitek et al.
Not to pick on one particular institution, but the difference between ‘free of charge’ and ‘open’ is important. We need open infrastructure to be truly resilient for the future. https://t.co/akenJsAiUA
Workshop: Post-selection Inference and Multiple Testing 7-9 Feb, Toulouse, F https://t.co/GpvGZI92n5
State-of-the-art collaborative editing for scientific manuscripts https://t.co/XJc4c07xbS
Great opportunity for starting up your own lab https://t.co/UxmmQBluqG
Postdoc position in Computational Systems Genetics in Heidelberg, with @Boutroslab and @wolfgangkhuber. Large-scale mapping of genetic interaction networks in cancer, combinatorial CRISPR/Cas9 and shRNA. https://t.co/1AGW2SW9af
Good meme, although not quite fair since some meetings and emails are about science. Defending uninterrupted time for concentrating on something is a constant struggle, though. https://t.co/1kNsqiCXCj
Research group leader positions in a large & vibrant institute - open call https://t.co/56wzLIRhOk
Found a new German word: R-ziehung
Workshop: Computational Aspects of Simulation and Inference for Stochastic Processes and the YUIMA Project https://t.co/V8PDYos8HN
The hills around @EMBL Heidelberg are still in the grip ot icy winter air. https://t.co/UWgWuS0JNz
Those who are funding science (and its dissemination) are not getting a good deal. https://t.co/16HnIEFWJH
Register now Summer School CSAMA 2018 Statistical Data Analysis for Genome Scale Biology Brixen, South Tyrol, 8-13 July https://t.co/yvQU5ibn0k
VJ Carey, Robert Gentleman, Laleh Haghverdi, W Huber, Mike Love, Martin Morgan, Johannes Rainer, Charlotte Soneson, Levi Waldron https://t.co/AZd2KmctRl
@ewanbirney On short term, maybe yes. But it’s not a null-sum game. In the end everyone is worse off with isolationism.
“Things spread through social networks because they are appealing, not because they are true. One way to make news appealing is to make it novel…” https://t.co/LQt6ImQCi3 - does this also apply to glam journals? (by @DrAnneCarpenter)
A tweet was posted from my account yesterday that I have nothing to do with. Account hacked? I found nothing about this through Google… https://t.co/Uo9Mp4riyF
Bioinformatics Engineer / Data Analyst Staff Position at EMBL (Heidelberg) Sequencing and mass-spec-based technologies to identify and functionally characterize RNA-binding proteins and their targets https://t.co/rWSmq9lJ8L
There are so many good collaborative editing platforms now: - Googledocs - Overleaf - Github/Markdown/LaTeX/Emacs Anythings is better than .docx email attachments… https://t.co/U7oleWflf9
Bioc2018 Toronto: call for abstracts, workshop syllabi, special interest groups https://t.co/q1JvFQCm6L
JuliaCall for Seamless Integration of R and Julia https://t.co/yGzqFls8Vk
R 3.4.4 is released https://t.co/8Kzqn4wkY7
@cwcyau A preprint is like a poster presentation. It’s not peer-reviewed.Treating it as such would undermine the whole raison d’être of the journal in question and its peer-review ethos.
You can try that with the editor. Sorry to hear this - it’s insane.
MacBook stuck with unhelpful error msg and dysfunctional after attempted update to macOS 10.13.4. Advice - wait a few days until Apple have sorted it out…
“[auth] failed to write file <private>”
JJ Allaire slides on R and tensorflow: https://t.co/EkQOtx65CQ
MDM4 is an essential disease driver targeted by 1q gain in Burkitt lymphoma - elegant work identifying genotype-specific dependencies of a tumor entity by HT RNAi. https://t.co/yMbs2P8d5C
This seems like an interesting project: description and re-implementation of the WhatsApp Web API, with the aim of writing custom clients. https://t.co/yZeTGJAvGL
RNAi screen finds potentially targetable vulnerability of mutp53 lymphoma “TRRAP is essential for regulating the accumulation of mutant and wild-type p53 in lymphoma” https://t.co/XKlFMPNBU1 Also, congratulations to Thorsten Zenz for getting this year’s Paul-Martini Award!
This is an interesting effort: a shared (R, Python, Julia, …) data science runtime based on the Apache Arrow columnar format. Blog post by JJ Allaire (@fly_upside_down): https://t.co/231smK53lu
Machine and deep learning for biological image analyis - Anna Kreshuk joins @embl https://t.co/Wpg5ZCto0Y
Protein stability and solubility dynamics during cell cycle - Great collaborative project by @embl scientists on https://t.co/DtVNZOoitd .Back-to-back paper by Nordlund lab: https://t.co/ZMFx74e9RY Via @nils_kurzawa
One choice of workplace for exploring latent factors and manifold embeddings at HCA-CZI collaborative computational tools workshop https://t.co/WY5DdT20Am
Easy access to a small collection of benchmark datasets for methods development in single cell data analysis for @humancellatlas etc.: https://t.co/m5NpDCZfci
The first single-cell sequencing datasets from the Human Cell Atlas are now available to the research community: https://t.co/Fubs7muvY8
After an SFO-FRA flight, it’s good to enjoy th cycling and the spring forest in Heidelberg. https://t.co/RsXqBiOHu1
Brand new ‘lfcShrink’ function that gives better (i.e. less stubborn) shrinkage. Try the ‘type’ argument: normal, apeglm, ashr. https://t.co/GBfjEIVw0u
Covariate powered cross-weighted multiple testing with false discovery rate control Completely rewritten version on arXiv https://t.co/jnGjxv7eHk This is the mathematical background to the IHW method https://t.co/0c7Zh7zvEV It’s been a real pleasure to work with @nikosIgnatiadis https://t.co/xXAJbnwGys
EU offers teenagers free InterRail pass - great way to explore Europe https://t.co/Kc3sL9yBBo For any EU citizens who turn 18 before July
New position as software developer in machine learning for bioimage analysis with Anna Kreshuk @EMBL https://t.co/K2Q22iUTuA
Interested in postdoc in statistical computing & drug-genotype interactions in cancer using exciting new data types? Consider EMBL EIPOD programme https://t.co/9vKrcAOWnc
More info: https://t.co/XZHQdHN6xf & https://t.co/LFUOSqC36d - now single cell, larger cohorts, combinat’x
- Go to https://t.co/v2NNAr2A9i
- git clone https://t.co/6yINjd8dvT
- On Mac, use the Font Book app to import the ttf files
- https://t.co/vCGe31iW9l
- Enjoy. https://t.co/oAEWQOU5o6
Many bioinformatic method developers think of their current project as the “final” tool in someone’s analysis, who will just look at the results. Au contraire: if a tool is actually useful, it will soon just be an intermediate step & other tools will want to import its output.
Indeed - one of the mighty ideas of the tidyverse is that of homeomorphisms: input class (dataframe) == output class. https://t.co/e1mimhljNr
@tammylarmstrong @JennyBryan Platonic ideas or Forms - (https://t.co/zNMut2ZWAg)
@ewanbirney Bayesian approaches become a lot more practical once you fit not only one, but thousands of models (e.g. genomics). Then you can use the data from the ‘all the other genes’ (loci, etc) inform the prior for the one you’re currently modeling. Empirical Bayes.
@ewanbirney Still one of the best explanations ever: https://t.co/EKWeP4FZNC Regular Bayesian: knows astrophysics Empirical Bayesian: just has seen the machine beep many times before
Munroe’s comment is perfect. Arguably there are no ‘statistical questions’… if you define statistics as making rational decisions based on uncertain data, it’s an interdisciplinary art that includes natural sciences, economics, history, psychology, etc. - besides applied maths. https://t.co/RwqwTx78w9
@Nick_Goldman @ewanbirney @aylwyn_scally I pointed to this mostly for the empirical vs regular Bayes contrast. It is however fascinating how many emotions the frequentist vs Bayesian one still stirs up.
Interesting package if you’re into bibliometry and data science https://t.co/9F18MyjjQ0
Want to know how the GDPR came about? ‘Democracy’ is a (surprisingly) captivating movie about the process: https://t.co/rav9CyQnnZ
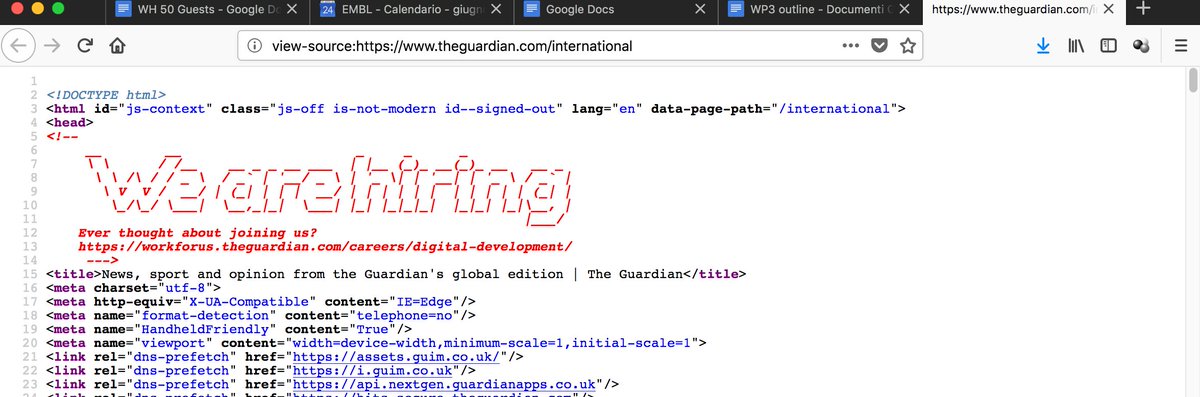
You get to see a job ad when you look at the Guardian’s HTML source code https://t.co/e2gxT81QLW
@vallens and to compare this to using Euclidean distance on the values after applying a variance-stabilizing transformation for the fitted NB noise model (such as from DESeq2). My prediction: should be similar.
@vallens Can you elaborate “don’t seem to work well for low counts”? VST is approximately the sqrt-function for low counts.
Thanks for the references, hadn’t seen them before. Indeed an interesting space, worth more work.
As you say, more generally this leads to feature (gene) selection or weighting - average count or variance is is likely to be only one the criteria that make a difference. https://t.co/YtYaz8zvkn
Agree. And it’s even worse than that. Some visualizations also impose structures, even when there are none. Incl. PCA/MDS plots, dendrograms, t-SNE. See e.g. https://t.co/KdMyEzLcDm https://t.co/E7S0vSV1S4
BioC 2018: Where Software and Biology Connect 26-27 July 2018, and Developer Day on 25 July Victoria University, Toronto, Canada https://t.co/OaAgjYcYzL
Manuscript authors: prepare figures within text at appropriate position, each legend with its figure. Having these all separate annoys your reviewers. And journals also prefer the more readable organization, e.g. https://t.co/jYwkAPFeYG
Imagine your reviewer not doing this on a 50” monitor but squeezed into a plane seat…
@SherlockpHolmes @AedinCulhane I wrote up (draft) some more explorations and visualisations on this: https://t.co/cpHuQqnQxM
@SherlockpHolmes @ZaminIqbal @MicroWavesSci @conTAMInatedsci @MBLScience I get a curious pleasure from reading the book on a mobile phone (being used to such things coming in PDFs usually)
@ewanbirney A main aim of collaborative repositories such as @Bioconductor is to structure and ease that load on individual developers.
@olgavitek @ewanbirney @Bioconductor - Can you elaborate your dependency problem? These should take way plumbing work from you so you can focus on content. If not so, needs to be fixed. - Maintainers do the support & training. User forum, releases, vignettes, workflows are meant to structure and ease this.
.@Bioconductor combines many different, distributed and strong-willed contributors. Each attempt at top-down standardization has a cost and needs to be weighed against letting diversity flower. https://t.co/gqjRu7Bzxe
@ivivek87 @emblebi Any matrix-like data. In the paper we include ex-vivo drug response phenotypes of the tumor samples.
@PlantEvolution Demand for ‘routine’ reviewing could be satisfied by a class of scientists who specialize in reviewing (and are rewarded for it).
And surely, who bothers to review would be a measure of ‘impact’.
@SherlockpHolmes @ZaminIqbal @MicroWavesSci @conTAMInatedsci @MBLScience Now also here: https://t.co/vuwCGe1G0y https://t.co/IjkdLtYAfX
@OliverStegle The Novelty (as a Westerner) of seeing the American president calling for destruction of your homeland: https://t.co/WFJoiVf44p
This is impressive: Human Rights Data Analysis Group https://t.co/wXjejuyWGD Statisticians can play important roles in media, think tanks, policy. Some exciting career options in these areas.
@lgatt0 @deDuveInstitute @UCLouvain_be Congratulations!
South Tyrol - where the vending machines sell organic cheese from the nearby farm https://t.co/V2Qspwv9fP
R is not end in itself - it supports statistical reasoning, just as well as paper-and-pencil calculations do. Also the approach taken in MSMB https://t.co/wljV9pioMX https://t.co/Mf1av3Reed https://t.co/5SOn01mRtp
DEAL-Elsevier negotiations suspended. “Elsevier demands unacceptable for the academic community” https://t.co/H1LVWwXwAc
‘Covariate powered cross-weighted multiple testing’ - the mathematical background for Independent Hypothesis Weighting https://t.co/jnGjxv7eHk v3: rewritten to be shorter & more concise
@MagnusRattray Yes.
@ksuhre Thanks - the IHW package is in @Bioconductor and easy to use, please give it a try.
In writing a paper, stick to one term per concept and to one concept per term. And as few as possible of them.
Why? Imagine reading a novel with a complex plot, lots of characters, and then these keep changing their names or identities…
@NimwegenLab @jepusto I’ve enjoyed Dostoyevsky a lot, although nowadays I prefer the writing styles of Orwell or Vonnegut, and they also seem more pertinent.
After a week of #csama and @Bioconductor, enjoying a bikeride to Pfannspitzhuette and the gorgeous mountains around Brixen https://t.co/LVj81leFgt
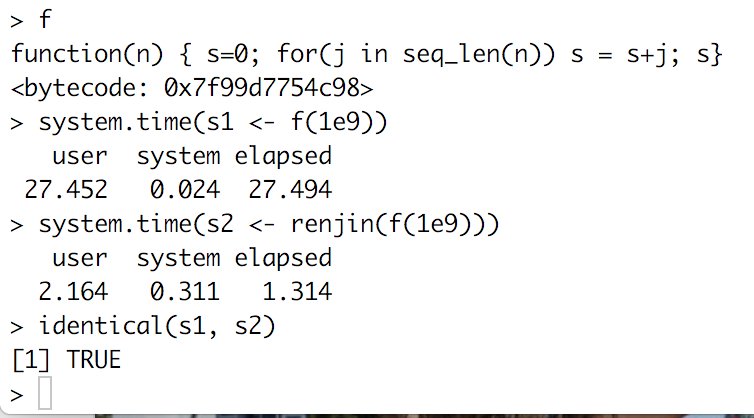
Made a small demo of the renjin R package, which lets you speed up R code by sending it to the JVM, and compared it with other options incl R’s bytecode compiler, Rcpp and vectorization. https://t.co/kyIL6RrVn4 @bedatadriven
@dselivanov_ @bedatadriven Happy to take your pull request.
Group leader position in bioinformatics or computational biology at Institut Curie in Saint-Cloud https://t.co/S9v55wCCUw
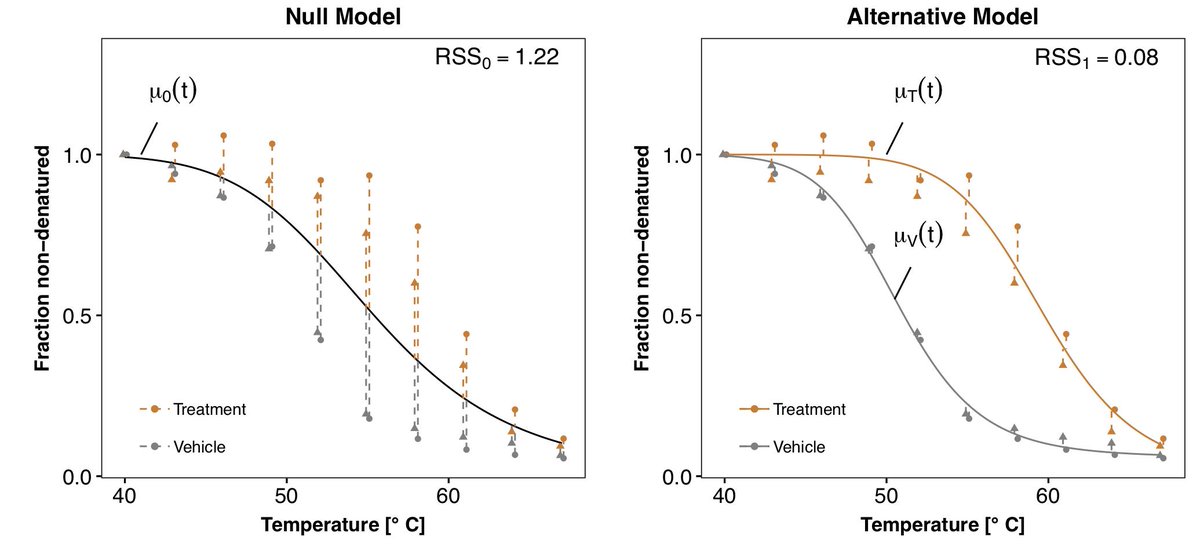
Non-Parametric Analysis of Thermal Proteome Profiles Reveals Novel Drug-Binding Proteins: https://t.co/6DyYPT06bK Gist: testing for effects of perturbations directly on the (smoothed) response curves is better than using scalar summary statistics like T_1/2 (or IC50…)
Join us at the workshop “New Directions in Single Cell Analysis” in Heidelberg 28-29 August Excellent speaker line-up Tutorials by @BrittaVelten, Aaron Lun, Simon @s_anders_m https://t.co/jQ5OQqZlAT with @OliverStegle Niko Beerenwinkel & John Marioni
This is a one-sided and polemic take. It lists imaginary risks of pre-peer-review release - but not real ones the gate-keeper model: the Wakefield MMR paper was peer-reviewed in Lancet, and it often denies important results from the public for years. https://t.co/iryJbSBDVn
If things were decided by rational arguments, the world would be quite a different place. https://t.co/VXz5kExXNV
Meet the @Bioconductor core team Two of them - @grimbough and @andrzejkoles are / have been based at @embl. #bioc2018 https://t.co/2AehBZfYeF https://t.co/bpt9kqyl6K
@ewanbirney @Nick_Goldman It links to https://t.co/PIw4voePWS https://t.co/BBgHAQOj0R
Brenda Andrews at #Bioc2018 on her amazing work on high-throughput imaging-based phenotyping of single and double mutations https://t.co/CcYHGab9wI
@PavelTomancak Victoria University, University of Toronto https://t.co/J9HruIvDCH
Comprehensive (meta-)tool for single cell RNA-seq analysis by the Harvard Chan Bioinformatics Core https://t.co/DxiPz9aiK3
I like it when talking to colleagues who work/-ed as director of >1000-staff institutes and they write their own code and care about ‘nerdy’ computational or statistical questions.
Thunderstorm closing in on Heidelberg after a hot summer day, viewed from mount Königsstuhl. @EMBL HD takes care of its employees’ well-being by lying in the middle of a network of beautiful biking trails. https://t.co/7wVd7DY2sS
When did you last time spot an instance of cargo cult science? https://t.co/mFuzGBQ1b2
The earth is now round in Google maps. Greenland no longer as big as Africa. @googlemaps https://t.co/tCc0505I1l
@markowetzlab Science is also about facts. In physics, only a handful of ‘natural constants’, but the more complex the systems, the more facts there are. Much of biology is historical happenstance, not logical necessity.
Sure. That original tweet was fanciful - but it had a point: the other extreme, where all the recognition goes to story-telling in 7-page PDFs, is in fact reality, and almost just as bad. https://t.co/M0D8RynHZX
Mixed wet/dry PhD position on tissue regeneration in top-notch place (Hubrecht) with one of the nicest PIs I know: https://t.co/Lm12YtALnu
@slinnarsson The mapping and modelling of structures in the data (‘clustering’) is bespoke and painstaking, a wild mix of biological priors and algorithms. Chapeau for being clear how complex and (from a computational / theory PoV) immature this still is.
Lot’s of good points here: “journals reject papers that use more rigorous methods than the discipline is accustomed to, simply because [they] are unfamiliar. Conversely, some disciplines become enthralled with methodology du jour without careful vetting.” “Cargo-cult statistics” https://t.co/xiY3w8TYuT
Ooops, “Lots” has no apostrophe. Wish Twitter allowed fixing typos.
@JEFworks @vallens @PetukhovViktor How about geom_hex?
@mikelove @SherlockpHolmes @thaasophobia @gilbertjacka @bhaibeka That’s indeed a trouble with ranks. Sometimes a better option is finding the ‘right’ transformation for the covariate. If it is itself a measurement (with replication), could aim for variance stabilization.
PhD position in evolution of animal cell types using whole-body single-cell RNA-seq, with Detlev Arendt, in Europe-wide project EvoCELL. https://t.co/nqjJZIu3LU
@davisjmcc @FertigLab Bioconductor experiment data packages: https://t.co/PootWlR2Nm E.g. Hiiragi2013, DmelSGI, HD2013SGI, RNAinteractMAPK, davidTiling
Schluechtsee is a little gem of a swimming lake in the Black Forest at 900m and only a short walk from the Rothaus state brewery https://t.co/ibcArhYy8a https://t.co/0dusj0TMbw
@daniela_witten The DESeq paper by Simon Anders & me was rejected by ISMB. Went on to Genome Biology, now 1000s citations.
@KingaKGdula Not at all unbelievable. Even if we agree that peer-review has a positive effect on average, there is a lot of stochasticity in it.
Especially judgements of “novelty” have often little to do with the manuscript and everything with the reviewer….
@rdpeng No, that’s a false dichotomy. Many scientific discoveries started from observations of nature (often, with new instruments). The questions came when someone looked at the data and said “that looks funny”.
The economics of academic book publishing are staggering. The “added value” by publishers comes at high markup; authors’ time and effort are taken for granted (given by them, perhaps tolerated by their employer). https://t.co/VjIynsgEyh
The system is ripe for disruption. I like the approach taken by @hadleywickham.
Major update of the rhdf5 R package by @grimbough - now supports HDF5 version 1.10 https://t.co/KcJjcVNULi
Inspiring line-up of topics and speakers at “New Directions in Single Cell Analysis” workshop https://t.co/jQ5OQqZlAT today and tomorrow in Heidelberg
Triple-omics single-cell analysis of mammalian gastrulation by Wolf Reik lab and collaborators - https://t.co/7REVVrgDN1
First talk today at the New Directions in Single Cell Analysis workshop
“Schools cede the task of evaluating PIs and their research to funding agencies”
And more of the risks of doing research is put on the individual researchers, esp the most vulnerable ones. https://t.co/ilgWYi7F5Y
Has anyone looked at the CO2 impact of deep learning? And compared e.g. to logistic regression?
Two attractive research group leader (faculty) positions at EMBL-Heidelberg
Genomics technology development https://t.co/O7BwFc7yPP
Genome biology (regulation across all layers of the dogma, genes & drugs, …) https://t.co/98SLY2BFuT https://t.co/K3R2eunKaj
@davisjmcc Congratulations!
Some cool research on somatic mutations in human health and disease here: https://t.co/zx9ABakKZ6 https://t.co/Ao4Zmp3GMO
@ewanbirney @Chris_Evelo Indeed FDR is more (and better) than p-values, and allows incorporation of additional (or ‘prior’) information, in a principled manner. See e.g. https://t.co/7Mmc8uc9zd and papers in Nat Meth & arXiv
I have signed the Permanent European Union Citizenship initiative https://t.co/JQr8pixXVu .
In 1850, the trip across the Alps from Basel to Milan involved trains, a steam boat and a horse carriage up to the Gotthard pass at 2100m, and could be done in 49.5h if no connection was missed. It cost 34 then brand new SFr. Today, of course, it’s a casual 4h direct train. https://t.co/S28Pb6Mi49
I can only second that! Great environment to establish a research programme in supportive, bright environment. https://t.co/fAhWJUJoNT
European Science Night in Heidelberg and Mannheim 28 Sep, 15:00-midnight Art exhibitions, performances, panel discussions, movies, concerts, hands-on experiences, tours, talks For teens and adults! https://t.co/pWvd2BLLeU (English) https://t.co/FVGmD8ZW4v (German)
Full house at Mari Sepp’s talk on cell type evolution at the Meeting of the Heidelberg Single Cell Center https://t.co/8oEvgwle9y
Future trend (wish): more combination of journalism with statistical data analysis & quantitative reasoning.Interesting example (incl. Jupyter notebook) here: https://t.co/nUqIBaKAEt
Short is good. https://t.co/uLd6BB9PmE
New EMBL Director General Edith Heard: “One of the reasons to accept this job has been Brexit.” https://t.co/gMe91dZmDR - support international collaboration. (EMBL offers plenty other good reasons, too.)
@hadleywickham Evidence of impact. Remind readers that software can be (is?) an academic output just like prose text. State how their software has changed the way science is being done, or thought about, in their field. Evidence of peer review / reputation.
Some clever person is selling one of my open access papers as a paperback on Amazon for 350$. The free version appeared in Genome Biology, DOI 10.1186/s13059-014-0550-8 https://t.co/NpfX4A5z6q …
There is something weirdly mesmerizing about translating a grant proposal that was written in English into German, and making up old-fashioned really long words for all the technical terms.
@onertipaday So it could be someone who set this up to convert grant money into private money. Not good.
@ewanbirney Hochdurchsatzphänotypisierungsanalyse, nullinflationäres Mischungsverteilungsmodell, Dispersionsdämpfungsnäherungsmodell, Genontologieterminus, Parameteroptimierungsprozedur auf paralleler Prozessierungsarchitektur
Human Technopole, a new fundamental life sciences institute in Milan, Italy, is recruiting senior and junior research group leaders in computational biology and ’omics: https://t.co/m6FW81Xrjr https://t.co/eIG4SofWTu
Group Leader in Biostatistics and Systems Biology Department of Cancer Genetics Institute for Cancer Research, Oslo, Norway https://t.co/YkU2yfxgky https://t.co/zGUssohkBK
So true in boinformatics https://t.co/QE3I2w5jlI
Register for the European Bioconductor Meeting 2018 in Munich, Germany, on 6-7 December 2018. https://t.co/JR8LW0vmNR https://t.co/yLHC3pj8zW
Seems analogous to biology https://t.co/bm13Ed7TNz
High-throughput CRISPR and single-cell RNA-Seq to study the genetic perturbation profiles of cell types within organs in vivo - looking forward to this exciting collaboration. https://t.co/4heumBkPEO
@BabyAttachMode Here is how it works at @EMBL conferences: https://t.co/O7McgGi1dR - not a grant, but a highly subsidized and practical arrangement
Ascona Workshop 2019: 16-21June Statistical Challenges in Medical Data Science https://t.co/axLJw7WwKN Save the date! https://t.co/sYpLHMwgcA
If you’re in the Heidelberg/Frankfurt area - highly recommended https://t.co/JlmYePK6GU
Testing should be taught as multiple testing first, with single testing as a peculiar special case - rather than single testing first and multiple as an afterthought.
Starting with two hours quality time with a good paper in a quiet corner, no phone and internet, makes the rest of the day (emails, meetings) so much more bearable.
@ErichMSchwarz The false discovery rate is a lot more intuitive than a p-value, and often what scientists actually want.
(And sometimes erroneously interpret a p-value as if it were a fdr.)
My impression is that this is similar not only for programmers, but also comp-bio researchers. Creating and protecting space for concentrated work has become the big challenge of the modern workplace. https://t.co/hlpS4388eP
Application 2 in https://t.co/Q7GqGd5eGe is on proteomics data, with number of peptides as covariate. Other data types where power tends to be limiting in my experience: eQTL (GWAS), HiC. With RNA-Seq, indeed power is often abundant enough already with BH. https://t.co/V9z0kzsz7P
@leonidkruglyak @michaelhoffman @ewanbirney Here’s a heretic thought: the distinction between peer-reviewed and otherwise published isn’t as binary as some would have it.
Looking at the code of someone who doesn’t now functions and loops, and writes complex programs by copy-pasting code blocks and mutating them. I imagine this is how genomes work.
Cobgratulations, Laleh! Well deserved. https://t.co/2GhT8pydaV
Friday thought: The enlightenment, as I was taught in school, seemed something from a remote past, involving dead white males in 18th century costumes.
No. It is on-going, by no means done, and we all should be involved in it.
While I like many of the people you meet there, I could not stand the management of that establishment any more. https://t.co/gaVeDDMgLo
Or a manuscript manuscript_final.docx https://t.co/YcpjbQkaLs
“We generally recommend against the use of zero-inflation in modeling sequence count data” (https://t.co/KwMI3kQavI) In fact what has been called zero inflation in older scRNA-Seq data (no UMIs, many PCR duplicates) is better thought of as excessively high counts. https://t.co/FxqAqEmzRg
That’s the way to go. https://t.co/5v7zmEmsXr
Farewell event to Director General Iain Mattaj - welcome address by Patrick Cramer https://t.co/RBCM8hnu9u
I think @IlianAtanassov refers to https://t.co/zfTmMvYoXL . Like many statistical methods, this involves some maths and abstraction, sorry @pwilmarth . There are also other good methods, e.g. collaborators use isobarquant. https://t.co/G5gjDvUwwp
Excellent points on the over-simplistic concept of ‘scooping’, from the editor-in-chief of Cell Systems. (And yes of course there are different degrees of urgency and importance to scientific articles, but I doubt that our current method of assigning these is much good.) https://t.co/r2oKvpWWjw
Has anybody with large mail volume (>400/day) had much joy with Apple Mail recently? Thunderbird works well, but I also like Mail’s “VIP” feature. And I wonder whether Mail is so clunky b/c of clutter in my settings/caches or just because.
Exciting postdocs with @s_anders_m and Hellmust Augustin computational & single cell biology - vascular control mechanisms of organ function during health and disease, from basic research to clinical application. https://t.co/g6MRCoG1b8
@markowetzlab @OliverStegle And what’s wrong with that?
There are different genres of paper. Impact and originality are two largely independent dimensions, both can be useful
@markowetzlab @OliverStegle I prefer scientific progress over entertainment anytime. There is a need for good benchmarking of methods, and it’s generally more ‘unmet’ than that for more algorithms
Bernd Bischl on mlr3 at the European Bioconductor conference in Munich https://t.co/nGHTBIymzt
@FelixFrauhammer Yup. See also https://t.co/iQKEPngEmV
Statisticians are just really drab at naming things: https://t.co/wcQY8CIh3l
@timtriche @markowetzlab @pathogenomenick @GenomeBiology @tuuliel @danjgaffney @michaelhoffman @OliverStegle And so, “novelty” is an even more ambivalent concept in methods research than in discovery science. On the long run, it matters more what is best than what was first.
@timtriche @markowetzlab @pathogenomenick @GenomeBiology @tuuliel @danjgaffney @michaelhoffman @OliverStegle Do bioinformatics methods exist since the beginning of time, and we just “discover” them? Or are they creations of our minds? https://t.co/FmKUI2DjB7
@OliverStegle @markowetzlab @rfschwarz When you (rarely) see a paper from your field in a glamour journal: - sour b/c you don’t like it / could have done it better, or - glad since it raises the field’s visibility ?
Master thesis project between myself and Christoph Merten on computational single-cell RNAseq analysis in antibody discovery using microfluidics: https://t.co/z1kbktBgdG https://t.co/87DWaNTcBN
Open position: Principal Investigator in Statistical methodology for systems omics, MRC Biostatistics Unit at the University of Cambridge https://t.co/XJNbQDVFuQ https://t.co/abUj6LYxJI
@emblebi @ewanbirney Congratulations, Ewan!
@OliverStegle @cwcyau Even if on average people end up poorer, some will make a fortune or gain power. A recurrent pattern in history.
@mikelove @anshulkundaje Agree with @mikelove. But there is a range: from being brought in late on the cheap to fix a flawed dataset with great effort and little reward; to being part from the start, complementing excellent wet-work (“hardware”) with dry-work (“software”) that takes it to new heights.
@mikelove @anshulkundaje Ironically, the first type of project can lead to really good methods papers (e.g. on “normalization” or “batch effects”).
@brembs Using journal rank in recruitment processes may often have sprung out of desire to make them fairer and less nepotistic - but the instrument is not fit for purpose and we need to move beyond.
Staff researcher position in biostatistics / data science at @EMBL ! Consulting and teaching statistical data analysis in fundamental biology research. Exciting & diverse work environment, lots of independence to shape things. https://t.co/28w4OQS1qp https://t.co/YYwMx1NXys https://t.co/wKmTxIH2og
Join the German Conference on Bioinformatics (GCB) 16-19 Sep 2019 in Heidelberg “Precision Medicine-where Bioinformatics & Medical Informatics meet” https://t.co/oWoE1Kq9LJ
@brembs At @EMBLEBI via the Image Data Resource (https://t.co/KOLpjDq6o9), or at Biostudies (https://t.co/AdpIt3bBpy). For electron microscopy, there is the specialized EMPIAR (https://t.co/PhVwYPsaAJ). Best to ask them for advice since it’s a moving space.
Scientists note! More scientists in the EU Commission would be a good thing. I.e., people trained and thinking as scientists; and not only in the EUC but in politics more generally. https://t.co/9LijzmpPaN
Registration is open for the 2019 Ascona Workshop “Statistical Challenges in Medical Data Science”, at Monte Verità, Ascona, CH,16-21 June 2019. Registration deadline: 15 Feb, https://t.co/xmSh3RGsui
Conference webpage https://t.co/axLJw7WwKN https://t.co/EcL481ZpQB
Who else is looking at someone’s pubpeer & retractionwatch records (carefully and reasonably) when reviewing their grant application?
Study on EMBO fellowship outcomes suggests new two-stage process: peer-review to select high-quality applications, then lottery to reduce these to available money. https://t.co/i8lN9yYM2w
I heard there was a lot of push back against this, as it admits panels decisions are not as good as they would claim, and removes power from them. Might initially also be a harder sell to funders (tax-paying public). https://t.co/MZR2u1930W
@OliverStegle On ICE from Frankfurt to Paris today. Average speed on German part of the trip (till Strasbourg): 117 km/h. French part: 281 km/h. (Peak is at 320 km/h). This is even if schedule is kept.
A German story (thread) https://t.co/LJ0tFH4y8z
Love this: “…this is a machine learning problem rather than a data science problem, because [..] the data […] doesn’t say which output value means ‘edible’ and which one means ‘inedible’.” https://t.co/ltpMWlXLZ2
The eco-fundamentalist farm shop in a village outside Frankfurt whose parking lot clogs up with Porsche Cayennes on a Saturday morning.
Staff researcher position @embl: develop methods for finding low-dimensional explanations in high-dimensional biological data (e.g. single cell multi-omics): latent spaces, graphs, manifolds. Important software eng. component. https://t.co/nJUaytHyWW Collab with @OliverStegle https://t.co/rnwkMRX0In
Do you think it’s rare b/c lack of calls, or lack of uptake by researchers? My impression is the second. Btw, EU also funded this: https://t.co/hutI9RMuko (~3 M€) https://t.co/AMuWJu2mXd
I updated a blog post about spurious structures in low-dimensional embeddings of distance matrices https://t.co/2hmCryROly
@lawrennd @Twitter I liked this tweet.
@ProfBootyPhD @aaronquinlan Yes, it’s coming out soon: https://t.co/bHDloxtgxY See also https://t.co/jSNISgPgBo - I hope the date for US shipping can be moved forward; or order from UK if you want it quickly.
Btw, the book is paperback only, for GBP 49.99, $64.99, €60.29.
Offers for hardcover on amazon are scams - I’ve raised that with amazon. (https://t.co/Ti7RDf6dM8 ?) https://t.co/lt1OqiYGzo
Two important contributions to the future of scientific publishing: “Publish first, curate later.” The current system of hard-to-get-in gatekeeper journals has become detrimental to science. Also see https://t.co/MH2DTVLfhS https://t.co/P0Nl0rT8DA
iphoto is so clumsy and unstable for photo sharing, am back to using scp/rsync.
@laurastephen @OliverStegle Yes, Dropbox works well for getting photos off phones. I’m interested in merging (selected) photos from multiple phones & owners into galleries.
@laurastephen @OliverStegle Besides that: on an Android phone you have a shell, can type into it through a bluetooth keyboard, or use ssh and some other unix programs.
Indeed, very recommendable Got this (as audiobook) for the long-ish Frankfurt-Heidelberg commutes I have to do these days. https://t.co/WebOzLceSg
@rafalab Seems anecdotal. I know other anecdotes where the good statistician discovered something unexpected and important; or where they overcome the data analysis bottleneck in a project. On average, I think a good statistical collaborator increases your productivity.
@mikelove @nikosIgnatiadis Makes sense. It’s different from gene set testing, b/c there is still one test per gene, but the prior implicit in the FDR control by (weighted) BH would be adaptive. Have not tried this yet! Am curious about people’s experience.
Thanks @SherlockpHolmes! https://t.co/QbE1qHTXvL
The generic paper abstract 1. Why is it important? (1) 2. What is “it”? (1) 3. But what is the problem with it? (1) 4. What did you do about the problem? (1) 5. What was your result? (n) 6. What are some wider implications? (1) (in parentheses: number of sentences)
@slinnarsson So true. Several good soundbites in there as well: “In the modern academic environment professors act more like middle managers than monastics” “…bureaucracy, which, once established, inevitably consumes the time and attention of its subjects to justify its existence”
@alos_31 @SherlockpHolmes It is: https://t.co/b8213McetR
Bioconductor 2019 talk / workshop / scholarship submission deadline: 15 March. The conference will be in NYC, 24-27 June. https://t.co/NonZKIeoUC
Two talks by @hadleywickham in Heidelberg: 12 March at HITS (https://t.co/YjLZQtgy2O), 13 March at @EMBL (https://t.co/b8v2eIM687)
TFW everytime you edit a collaborator’s manuscript, you point them to ISO 80000-2 (https://t.co/NTkSRzkyai)
@pedrobeltrao True, but I am not sure “failure” alone cuts it. Some people are getting exactly what they wanted: destruction.
Crossing the Alps for a first visit to HumanTechnopole in Milano, Italy. https://t.co/6mSVMbgQLY
I want to KonMari committee memberships.
We invite you to this year’s edition of the summer course Statistical Data Analysis for Genome Scale Biology in beautiful Brixen, Italy, 21-26 July 2019. Course programme and more info are here: https://t.co/2cnkGAJpfX #csama Please retweet or forward to interested colleagues https://t.co/S0nsTCTVUv
Postdoc Position in Computational Single Cell Genetics / Statistical Modeling Large-scale Perturb-Seq in in vivo ERC-funded project in collaboration with O.Stegle, M.Boutros, J.Lohmann https://t.co/gH58yC0bvt https://t.co/vbMZxeo3eK
Thank you to @SherlockpHolmes for such a wonderful, and fun collaboration. https://t.co/ZV6FeVCwmR
Huber group @embl celebrating the joy of doing science, international women’s day, and Holly’s birthday in MainNizza https://t.co/mRTLCDZqNL
… the location of the VIZBI meeting next week: https://t.co/clDyFoHsBp
@dermitzakis That’s nice, but the bottleneck is transition into faculty. What is the fraction of women in your institute?
There’s also a livestream of H. Wickham’s talk today on “designing tools for data scientist-programmers” https://t.co/jwPX3Y7bef : 11:00 CET.
In case you cannot make it to the venue, beautiful Villa Bosch next to Heidelberg castle. https://t.co/s7znpXSKGO
The voice of reason (thread). https://t.co/wVKm7k7OnU
.@embl Summer School: Visualising Life For advanced undergrad students from non-biology courses (e.g. physics, engineering, mathematics, CS), exciting insights into interdisciplinarity of modern biology research 15-26 July 2019, Heidelberg https://t.co/Wk2nSMiE3s https://t.co/6kETJlwpji
Slides for my talk at VIZBI 2019: https://t.co/OapOYIc4qq On horseshoes and other artefacts in low-dim embeddings of high-dim data
Just discovered this marvel of an opinion piece by Albert Einstein from 1916, as pertinent today as then: https://t.co/x4KyGORZKl The struck through part seemed so controversial to the publisher that they had it deleted. Plus ça change… https://t.co/fjbM3WypZg
@alishum_ali @SherlockpHolmes Hope you enjoy it!
Great that you wrote this up. This is so evident if one actually looks at the data; or spends a few minutes thinking about the physics of the measurement process. Yet so much heat generated for fantasy models of ‘dropouts’. https://t.co/L8R2hMHiiN
This is useful. https://t.co/jXPG8iLcsx
Good news for the biological data science and ML research communities - and congratulations to @fabian_theis ! https://t.co/J7vIz5a1aZ
@slinnarsson @PetzoldKatja I’d stay away. The website has no impressum. Looks like an impostor under misleading flag trying to syphon off money from naive grant administrators for trivial or made-up services. Quick internet search leads to https://t.co/TzhKjO2yJy
@slinnarsson @PetzoldKatja Btw, I understand that some of the excessive-looking paperwork with EC grants is a reaction to past experience and future threat of fraud, across dozens of countries and legal systems. Not great, but hard to see how else to do it.
Little opinion piece on reporting p values, in particular, very small ones. (This is unrelated to the recent brouhaha about abandoning them altogether, or their dichotimization. Just about one basic sanity rule using an imperfect tool.) https://t.co/Ps8sIj6JWq @QuinceyJustman
@QuinceyJustman @SherlockpHolmes Works freely (i.e. no paywall) for me.
This grew out of a reply to a reviewer’s comments on our draft for https://t.co/9WgTqqteIz - reviewer didn’t like our “p < 2x10^-16” and wanted to see small numbers. @QuinceyJustman’s idea to turn into a “Best practices” piece.
Untested ctDNA tests for cancer, attention-seeking press release, allegations of IP theft, pushy commercial interests - story brewing at Univ. Hospital Heidelberg https://t.co/i9iBtL5cHC (in German)
@gavinpaulkelly Thank you, Gavin! That was indeed an oversight. I fixed the post https://t.co/2hmCryROly
A more literary version of this meme: “The Everyman -who yearns for a sense of shallow propriety as well as for a deeper sense of belonging, even if it comes at a great price, including that which is sensible or even necessary for his own survival.” https://t.co/rNl2cU6HPW https://t.co/IhVKvdLbdf
Btw, Max Frisch is one of my favourite authors, worth checking out.
The Event Horizon Telescope consortium published their black hole picture in The Astrophysical Journal Letters rather than Nature or Science. Unimaginable in biology. https://t.co/CkE3aQO9NV
@twittkopp Congratulations, And welcome to @EMBL!
Congratulations to Katharina Imkeller @K_Imkeller for the Promotion Award by the Society of Biochemistry and Molecular Biology!!! https://t.co/Kr9W1m0WyS
To my EU followers: please make sure you are registered for the parliament elections in May, and vote! See e.g. https://t.co/6bNZCuTybR
Analyzing shape changes in thermal proteome profiles with functional data analysis (instead of summarizing into “melting temperatures”) increases the scope of this exciting technology - substantially revised manuscript https://t.co/22WEhBaoFF
TED talks are not usually my thing, but this one by Carole Cadwalladr is remarkable: https://t.co/3vzhqhe0iT On the role of Facebook in undermining democracy
R 3.6.0 is released: https://t.co/9ktim9lzL4
@ewanbirney Good move. Susan’s and my attempt at giving a simple intro: https://t.co/CYOoCY9X9D
@florianjug R and EBImage.
Congratulations! https://t.co/GINrvMeAnW
@grimbough @SherlockpHolmes Thank you, @grimbough! Only a few cosmetic code updates were necessary this time, even though the book depends on some 460 packages. (Let’s see how that goes in the future. :)
Welcome to Twitter @kaessmannlab ! Groundbreaking studies (and data) on functional evolution of mammalian genomes and cell types. https://t.co/E3SLjkGvzQ
Job offer at EMBL (Heidelberg): Biostatistician for the Center for Statistical Data Analysis - to offer consulting services & training to scientists across the institute on biological data science. https://t.co/cacg5ijAOF https://t.co/wHc5hFXsyb
Postdoc in machine learning at @embl with @wolfgangkhuber: Multi-omics factor analysis - develop methods for finding low-dimensional explanations in high-dimensional biological data and apply to scientific discovery in biological and biomedical research https://t.co/nJUaytHyWW https://t.co/NJy6EAkXFC
@fabian_theis @jensspahn That seems an astute analysis.
@mikelove @Bioconductor @F1000Research The Reproducible Document Stack (RDS) project (eLife, Substance, Stencila) aims at an open-source solution to publish reproducible manuscripts through online journals https://t.co/hjMUHFazPw I’ve no direct experience of it (yet).
Professorship (W 3) for Computational Cancer Biology in Cologne, in the newly established Cancer Research Centre Cologne/Essen: “generate novel insights into cancer biology with the intention to improve the clinical management of patients” https://t.co/XE1A7Cso6D (Medicine)
TFW in every manuscript you proofread as a co-author, you need to educate everyone else on dangling participles https://t.co/5xy3r43RaS
@markowetzlab @robjohnnoble I’d use different language, but basically agree. Couple of red flags here. The job conditions don’t match the expectations, one of the two will have to give. Some places still seem to be struggling w seeing comp. biology as a proper & strategically important scientific discipline
@BenLehner @eu_comission @ERC_Research Network projects have been gamechanging in making researchers from different countries talk to each other and creating some sort of continent-wide scientific arena. Calling all rubbish is polemic & unfair. Quality can be certainly improved, and we need less of them and more ERC.
@mbeisen @eLife There’s probably a couple of subtypes here: 1. same data, same question(s), different method(s) → same or different conclusion 2. same data, new questions 3. multiple previously separate datasets, new questions Each is worthwhile. #1 perhaps the most sorely missing.
Cool idea. This promises to be really useful for high-throughput biological data. https://t.co/dqkebYGMsa
If you’re interested in setting up an interdisciplinary postdoctoral project on computational analysis and modelling of animal development and regeneration in a cool model system, with Aissam Ikmi and myself, please get in touch https://t.co/ISAt9w7tWU https://t.co/UoSMRFS3Ai
@tslumley Great detective work by @tslumley on a sensational result that wasn’t (“autism is an infectious disease via the microbiome”) and another reason why authors, editors, reviewers & readers should insist on publishing the code of such analyses, rather than vague verbal descriptions.
@tslumley And while I am not in a position to fully understand the ethical trade-offs involved, it seems that after the Wakefield disaster, responsible editors (of scientific and popular media alike) would tread more carefully. @TheEconomist
@michaelhoffman @OliverStegle @tslumley There are none. (But code is less useful w/o data.)
Unrelated to code, what I meant with “ethical trade-offs” in my previous post is about how careful to be before making an extraordinary finding public: neither too retentive nor too gung-ho.
Hypothesis: glamour status of a journal encourages (sometimes) brutish and nasty reviewing, compared to more constructive and careful approach available in specialized environments. (And maybe that’s fine - just how the world works.)
Opportunities for interdisciplinary postdocs at @EMBL. Contact us at https://t.co/KRwwdeNYF5 for projects in biomedical data science, statistical method development in ’omics & imaging, biological discovery. https://t.co/QumN8QztwW
Postdoc position in Computational Biology on Somatic Evolution and Early Detection with the fabulous Angela Goncalves and Duncan Odom - https://t.co/kKhTncw9KR
R 3.6.1. is released - various bugfixes and robustifications https://t.co/mnUhWsFnqs #rstats
@TheEconomist Given the flaws of the paper, the potential damage caused by parents panicking that autism might be infectious, and the history of suffering caused by the A.Wakefield scam, this is irresponsible reporting by @TheEconomist. * https://t.co/T6leNgcOLN https://t.co/qqVWDVzIrw
Asymmetry in before/after ratios in CRISPR screens: https://t.co/BqRqCqY3db by @K_Imkeller. Big implications for statistical hit detection and experimental design.
This asymmetry is a unique property of such experiments - it does not occur, e.g., in typical RNA-Seq. It means that DESeq2 or similar methods are suboptimal here. Ultimately, it is down to the irreversibility of time and the second law of thermodynamics.
@DrAnneCarpenter Good point. Will define in Abstract in next version.
@markowetzlab Well the asymmetry has to come from somewhere. Why is this situation different than, say, differential expression in RNA-seq. So when you trace it, that’s the place.
Lecturers who want to use Modern Statistics for Modern Biology (https://t.co/VeF6EmIFVy https://t.co/obiejJIy4R) in a teaching course can contact @SherlockpHolmes or myself for additional teaching material (slides, homework labs)
Team Leader Positions in Advanced Light Microscopy | Electron Microscopy Service and Technology Development at @EMBL Imaging Centre https://t.co/3wJEOIVw4Z https://t.co/Zdo6TvOkmD
Irrespective of their views and achievements (in which I see good and bad),
this is a historical picture. https://t.co/cXkbUdYRPq
Thoughtful contribution by phantastic group members to @embl Day. https://t.co/8uQneWk6l1
Makes me think of all those people working in the Institute of Batch Effects. https://t.co/8uQneWk6l1
Aim is to make DESeq2 runtime O(n) in number of samples (single cells), which it wasn’t in the past for no good reasons except that it disn’t matter then. https://t.co/7Wh99Gte8x
The resilience of genes in CRISPR-engineered KO cell lines: Biological Plasticity Rescues Target Activity in CRISPR Knockouts
https://t.co/lyYAYpcNtk – @FredOnion Arne Smits Cellzome/GSK @LarsMSteinmetz https://t.co/C7qLSQQJ3p
Quite a pattern these days of angry old white men hating politically active young women. I wonder who will have more staying power. 🦸♀️ https://t.co/pxO4iFfwvP
@mikelove @paulfharrison Also variance stabilizing transformation (as in DESeq2) is interesting if you want to feed into clustering, classification, visualiz. etc. But a fundamental problem with transformations in the presence of different (library) size factors is confounding effect size & significance.
Postdoc on computational single cell ’omics - high-throughput perturb-Seq in vivo, in adult organs, to understand multicellular tissue development and maintenance. Looking for statisticians & comp.-biologists. https://t.co/gH58yC0bvt - apply till 18 Aug https://t.co/KRwwdeNYF5 https://t.co/cXjS00Fb60
Germany most popular non-English speaking country for studying abroad, and 4-th overall (after US, UK, AU). https://t.co/ERIuWDPGRe https://t.co/MACg7FPXQC
Interesting in running a “Statistics Clinic” at EMBL? - Statistical advice and hands-on help on biological data analyses for researchers conducting cutting-edge molecular biology research - Computational statistics and data science in biology https://t.co/zPPktcaj5D https://t.co/Azzu3qv6w5
Machine Learning courses/workshops with Anna Kreshuk (@ilastik_team) and Bernd Bischl https://t.co/2YSzGrclKD
TFW when your ms got bloated with tangential analyses to satisfy reviewer requests and then after acceptance the editor asks you to shorten it to the most essential pieces again.
Almost all human activity is moving things from one place to another.
@GenevievMichaud “Eat your own dogfood” is an expressive term (also slightly gross), and not from me: https://t.co/jbKYj0Il4v It’s certainly my experience that bioinformatics software needs to be tested in real projects (messy, time-pressure, multiple aims etc.) and usually this changes a lot.
It’s 2019 and OUP wants me to pay £350/€525 for colour figures in printed paper versions of my article that they will then sell to libraries. Fallen out of time…
(Btw last time £/€=1.5 was in 2007.)
@emmamarydann @embl @UU_GSLS Thank you, Emma, for working with us. It’s been a pleasure. And so successful!
@shl Could you quickly run your pipeline over my data?
@HITStudies @RichBonneauNYU CET or CEST?
Edith Heard, the Director General of EMBL https://t.co/4lwEmxlrem
Alarming, and strange: https://t.co/mSrKVJJbZH
Strong team of scientists leading the buildup of the new Human Technopole research institute in Milan. https://t.co/DN8ErlYyJI
Oliver Kohlbacher on bionformatics tools and infrastructures for supporting precision medicine personalized vaccines at #gcb2019 https://t.co/0avkzIKTLl
Robert Gentleman setting up for the keynote talk at #gcb2019 https://t.co/JkUZuwrDZX
@FredOnion @RNASeqBlog @DanaFarber There are (by and large) no missing values in sc-RNAseq, and the concept of “imputation” is misguided. There is sparse sampling, and “smoothing” might be the right way to think about reducing noise in the data.
Congratulations, Holly! https://t.co/TENuh5eQRQ
Travelling FRA-SFO with our 9-months old. Just taking off. So far UA staff have been really good. And baby happy. Looking forward to teaching Bios221/Stats366 with @SherlockpHolmes at Standord. https://t.co/vuwCGe1G0y
This from Robert Harris…. https://t.co/zBRLFjxXWv
Thermal proteome profiling is a mass-spec based technology for detecting drug binding to proteins (or other modulators of protein stability) in living cells. Here’s a statistical method and R/Bioc package for sensitive and reliable analysis of such data. https://t.co/nUVDqQGWmU https://t.co/CXkHKV1QOh
Fill out @Bioconductor ’s first community survey to collect feedback on the project and help guide future activities: https://t.co/Jrm9i2leOW
Genomics Technology Development Faculty Position @embl Great place to set up your lab – excellent package (core funding, PhD student program, facilities) & nice colleagues. EMBL offers many career development opportunities & a collaborative atmosphere https://t.co/m9yRXyuSut
Just deleted Thunderbird incl. all personal configuration files & reinstalled from scratch, b/c after system update new messages weren’t bolded anymore (now they are). I guess that’s the equivalent of doing a full house-cleaning after discovering a few breadcrumbs under the sofa.
@MundAndreas @FredOnion @GSK @LarsMSteinmetz Cool. Another impressive example. The contribution of our paper, I hope, is the large-scale, systematic nature of the analysis of the phenomenon.
Insightful systematic study of CRISPR-KO efficiency with surprising results by @FredOnion, Arne Smits and GSK/Cellzome colleagues https://t.co/IZHGQl3vuh
An account that keeps reminding us what can happen if we allow the bitter, spiteful and hateful to run a government. https://t.co/0vPuRJzxEP
Congratulations, Nils! https://t.co/oQOVkjFod8
In my introductory statistics lecture on hypothesis testing, I teach FDR/fdr first and p-values second. It seems more logical and more natural.
@bbrsntd Discovery = rejection
@rafalab This is very relatable. Nevertheless, I prefer statistics playing the role of enabler over that of gatekeeper: enable scientists to discover real and interesting phenomena at the edge of detection limit, in sometimes messy and complex datasets.
Here the slides: https://t.co/crowFitsOV
Good that ‘Research’ is now explicitly mentioned in the remit of the new Commissioner. https://t.co/C7aPktWR8Y
@MarinaP63 This was the 1990s and I thought I would open a mountainbike rental shop on one of the Canary islands.
@tamas_schauer @QuinceyJustman No. See e.g. GWAS.
Congratulations, @Michael_Boutros ! Great news for science, and for @DKFZ. https://t.co/Bt3EVGF5Zt
Passing through there on my way to #bioceurope2019 in Brussels. https://t.co/aMRaB68n1K
#EuroBioc2019 conference dinner in the windmill farm near UC Louvain. Animated discussions on multi-omics, proteomics, single-cell, containers, reproducibility et al. https://t.co/MwjecmwpoN
Congratulations, Kiran + lab! Exciting times ahead. https://t.co/7Ep3dRVE39
A succinct summary. https://t.co/h4SSHMvXrk
Lunchtime ride in the hills around @EMBL on a bright and warm December day. https://t.co/OGCiS34qbz
On top of the hill, our neighbour, the Max Planck Institute for Astronomy with a futuristic building complex hidden in the forest. https://t.co/mja3uGQIdR
Changes by @ERC in its panel structure (from 2021 / Horizon Europe): re-balanced panel topics in life sciences and one more panel each in physical and social sciences/humanities: https://t.co/7IPsZwaLqe The panels: https://t.co/nvpyxrcITf
@erc The “application patterns” (i.e. numbers of applications) seem to have led to a slight increase in physical sciences and social sciences/humanities, whereas size of life sciences remains the same.
My favourite adjective is “good enough”. https://t.co/bh8sdAeQrj
An excellent initiative. Something that @EMBL will also enact more explicitly. (In my experience the spirit of DORA is already being followed in recruitments & promotions.) https://t.co/ssKdLCk0Eb
Without #Erasmus I would not have gone to Edinburgh as a physics student, would not have considered UK among places for postdoc, would not have applied in Cambridge (EMBL-EBI) for my first faculty position.
Another instance of today’s old screwing the young. https://t.co/j7Ra4Wij0K
@vallens @tangming2005 @adamgayoso This statement is not true. It is mixing up sufficient and necessary assumptions. Least sum of squares estimates can be unbiased, minimum-variance, etc. under many additional conditions. Ref: https://t.co/7uT23lqbG5 or just google “least squares normal distribution”
@vallens @tangming2005 @adamgayoso Many users of statistics worry far too much about normality assumptions and far too little about independence (a.k.a. batch effects, normalization,…)
@tangming2005 @adamgayoso @vallens For sc-RNAseq data, glmpca is the way forward: https://t.co/RKrqh5F3D9
@ewanbirney There have been plans for a direct London-Frankfurt train, which are on ice. Maybe can be revived in context of planned new green investments by Germany and EC… https://t.co/5WAMR7HJ3r
@lgatt0 @NicholasStrayer See also a head-on attempt at modelling the non-random missingness in “Probabilistic Dropout Analysis for Identifying Differentially Abundant Proteins in Label-Free Mass Spectrometry” https://t.co/V1yycJCEjw
@raphg @davisjmcc What do you even want to impute? Zero is a data point, not a missing value.
However, sometimes (esp. in CS circles) I’ve seen the term ‘imputation’ used for a goal that statisticians would call ‘smoothing’. And that does has merits.
@lgatt0 @NicholasStrayer By @const_ae and @s_anders_m
@MagnusRattray @daweonline @tangming2005 vsn works well for continuous data, see package and assoc. papers. For count data (Gamma-Poisson), a different but similar function can be useful as long as the majority of action in the data is for higher counts. Otherwise, discreteness leads to artefacts (as it does for log).
@ewanbirney @e_petsalaki Wise words, Ewan, thanks for stating them.
I had to drop out this week out of EMBL’s semi-annual PhD student recruitment round (a big deal) due to sickness in the family and babycare. Apologies to students and colleagues who were affected, but there was no other way.
Say what you mean and mean what you say. No synonyms. One word, one concept. https://t.co/PLpj97Jdal
Equally true for bioinformatics. https://t.co/TihJ5d1eXb
Two lessons I’ve drawn out of this: 1. Good experimental collaborators are essential. They are also hard to find. Once you find one, hold on to them. (Corollary: at least as important as their scientific brilliance is that they are good people.) (2/3)
- Bad datasets tend to take more time to work with than good ones. This can easily lead to a very bad bottomline on your productivity. Monitor cost-benefit and cut projects accordingly. There is more data, and more ideas, than time.
Postdoc in my group @EMBL for biostatistician / bioinformatician to develop methods and co-design & analyse new datasets on cancer recurrence using proteome and metabolome mass spectrometry & other ’omics https://t.co/82NKb2rdsc
Finite sample results on Independent Hypothesis Weighting (IHW) for covariate powered cross-weighted multiple testing, with @nikosIgnatiadis: https://t.co/jnGjxv7eHk (1/2)
@nikosIgnatiadis The Benjamini-Hochberg method is simple and beautiful, but has suboptimal power when tests are heterogeneous - which is almost always. IHW lets you use covariates that may be associated with each test’s power or prior probability to improve overall power https://t.co/0c7Zh7zvEV
@DarwinAwdWinner @FedeBioinfo @mikelove @nikosIgnatiadis @keegankorthauer Here’s @keegankorthauer ’s paper: https://t.co/xHbBhsnrlk https://t.co/tZP4LotQAN
@amuellerml There is a microbiome and a gene expression example with lasso in the Modern Statistics for Modern Biology book by @SherlockpHolmes and myself: https://t.co/H1MY82kMiJ
@lawrennd @amuellerml @SherlockpHolmes @marta_milo Just to expand on Neil’s reply: https://t.co/61yQpN8iWq
And indeed https://t.co/irDSsZLoXl (or RNA-seq).
Lasso and ridge regression for ’omics data have been very widely researched since the earliest days.
@denbiOffice CSAMA 2020 in Brixen/Bressanone: course website and registration are open https://t.co/84ABHeaxWG
CSAMA 2020 in Brixen/Bressanone https://t.co/MCZ9l0S4Q5 Summer School on Statistical Data Analysis for Genome-Scale Biology, with @Bioconductor
@K_Imkeller @BrittaVelten @lgatt0 @grimbough @mt_morgan @LeviWaldron1 @CSoneson LoriShepherd R.Gentleman @drisso1893 @jo_rainer S.Bell https://t.co/NqdjQoMpug
Highly recommended: https://t.co/V1DzkIambH
Supposed to write one of those research productivity reports for my institute, which means it is high time for a complete refactoring of my latex and bibtex style files.
@erikacule For the publication database behind e.g. https://t.co/7Lju3OZet4 I use a combination of bibtex and R scripts (with @grimbough) - a complex and hard-to-maintain tool chain, but I find it hard to think of a better alternative. Ideas?
@OliverStegle Debugging code is more exciting than writing elegies about oneself.
Bioconductor conference 2020, 29-31 July, Boston. https://t.co/payJBak0qN
@Alfons_Valencia @jonsv89 @juanrrivas As a recruiter, I think it’s important the student made good use of the time and resources they had, and that they come with the relevant skills. In other words, speed = distance traveled / time, and this depends both on numerator and denominator.
@VeraPancaldi @Alfons_Valencia @rionbr @jonsv89 @juanrrivas It’s a two-way street. Institutions that insist on mono-culture and linearity will in the end be less competitive than those who embrace diversity. As people live longer, and science is changing rapidly, we need more ways for retraining and non-linear careers, not less.
@nils_kurzawa @UniHeidelberg I love LaTeX, but it’s been plateauing for almost 20 years now, and does not easily render into HTML. Rmarkdown offers an alternative authoring platform that is more modern and renders into attractive HTML pages, PDF, and other formats.
@nils_kurzawa @UniHeidelberg We wrote the book https://t.co/obiejJIy4R with LaTeX (@SherlockpHolmes), but I wish it were Rmarkdown (and see no real reason why not)
@davidmasp @nils_kurzawa @UniHeidelberg pandoc is the engine and Rmarkdown is the authoring environment. Both are heroes.
@johnpoverington Aren’t all proteins related once you go back far enough in time? Probably structural or functional (dis)similarity is a more useful concept.
@_richard_feder @carlesgelada @lawrennd Different fields seem to put different amounts of incentives on proof-of-concept vs implementation, and I suspect those who value implementation more produce more impactful science and more real advances. “Standing on the shoulders of giants” requires quality shoulders.
@InesBarroso4 I’m sure everyone who gets an individuals-oriented price has well-deserved it. But it does seem a bit creepy to me to put so much attention to personal fame & glory when most scientists are in it for the joy of knowledge.
@inespinedatorra @NPirastu @InesBarroso4 Doing science is more like building a cathedral than like painting the Mona Lisa…
Surreal but probably to the point. Travel plans, big meetings, … https://t.co/CA7FLfsJvz
Two of my favorite science anecdotes are how Schroedinger and Heisenberg each had their breakthrough ideas in quantum mechanics. Not sitting at their desks. Heisenberg, while hiking on the small North Sea island of Helgoland, where he had escaped from an attack of hayfever (1/2)
in his workplace Goettingen. Schroedinger, on a skiing holiday in the Alps. Of course both also worked hard in regular office settings and had many meetings, in order to even be able to have such ideas. Nevertheless, if you’re a scientist, isn’t it a good excuse to get out more?
@hjpimentel @PhDemetri Not necessarily, if you have lots of models and lots of associated p-values, there may be a way out - look at @nikosIgnatiadis work on cross-weighting https://t.co/K9D7t3yAGU
Interesting work by @K_Imkeller showing a role of the second law of thermodynamics in CRISPR screen analysis. https://t.co/nNf34I9VYa
Supervised cell type assignment from sc-RNAseq data: https://t.co/jXvVwe2LUd “clustering-based methods … can artificially identify more cell types as the dataset size increases, without reflecting biological ground truth [and] depend on arbitrary user decisions”
Nice work in similar vein by @FelixFrauhammer presented at Heidelberg Single Cell Center Meeting using smoothing and Gamma mixture models.
Only worry: what to do with all this creativity and energy people like to spend on Rohrschachplotting t-SNE plot clusters…?
@dermitzakis I am thinking of my grandmother Margarete Hertel (1906-85), who as a single mother & war widow brought up four little children during and after the war, through poverty, hunger, social exclusion as a migrant and surrounding brutality. And was a generous and loving person.
Professorship in Lyon https://t.co/xKjMbhrC7a https://t.co/7yUL7nB9uW
While everyone is setting up for extended stays at home, here’s an amazing piece of electronic music history: https://t.co/7RCKLj6rzg by @tomita_isao1932
One of the best pianists https://t.co/LTxBxNuSLW
Staying at home with my wife and 15-months old child. There are lots of issues, of course, not all fun. But one of the more intriguing ones is our massively increased consumption of chocolate and champagne.
@dharap Luckily, solo running or cycling is currently allowed in Germany (and in fact encouraged, since it’s good for your immune system, your lungs, and your sanity), and the weather is nice.
For everyone who is now upping their bioinformatics “best practices” game, a reminder of Jenny Bryan’s (et al.) Happy Git and GitHub for the useR page: https://t.co/ust4VnC5zt
. Should’ve tagged @JennyBryan - here we go.
I sublimated some frustration over poor internet connection in the home office into a script that continuously monitors quality parameters. It is based on a command line interface to the speedtest service, crontab and visualization in R: https://t.co/GjLOlgSwVG https://t.co/ejp4pClNzG
Statistical analysis for omics pet pieve: do not use “patients” and “patient samples” as synonyms. There can be multiple samples per patient (across time and space), and patients are people.
Or @bioconductor if it’s about biological data and you want to additionally benefit from Bioconductor’s data structures, I/O, helper packages, and support infrastructure. https://t.co/NsU64VVozi
R 4.0.0 has been released. What a milestone. Thank you to all the contributors in R-core and beyond. https://t.co/H7n9WVwL7z Several significant user-visible changes of (previously confusing) behaviour, and many new features. https://t.co/7W5We8Xsng
@olgavitek @cendrinou @drisso1893 @stephaniehicks @drighelli @ftoscanomd @mikelove @fannyperraudeau @koenvdberge_Be @p_boileau @cazencott @JennyBryan @ljacob @SherlockpHolmes @BhramarBioStat @AedinCulhane @pedjagogue @Gundrylab Be kind. To others and yourself.
Six-word story challenge in #Covid19 - nominating @areyesq @CSoneson @FedeBioinfo @markrobinsonca @s_anders_m @mt_morgan
@olgavitek @cendrinou @drisso1893 @stephaniehicks @drighelli @ftoscanomd @mikelove @fannyperraudeau @koenvdberge_Be @p_boileau @cazencott @JennyBryan @ljacob @SherlockpHolmes @BhramarBioStat @AedinCulhane @pedjagogue @Gundrylab @areyesq @CSoneson @FedeBioinfo @markrobinsonca @s_anders_m @mt_morgan My close contender was
Babyfeeding nappies zoom zoom cycling chocolate.
@LomascoloSilvia @pdalgd @KorichevaLab @SherlockpHolmes Most authors will be happy to see their ideas and work get out into the world. Online is so much more accessible than a physical book (and royalties for textbook authors mostly are minuscule compared to the effort) - this is why the book by @SherlockpHolmes and me is free online.
@LomascoloSilvia @pdalgd @KorichevaLab @SherlockpHolmes …see https://t.co/Qjlp7WmCf0 https://t.co/obiejJIy4R https://t.co/VeF6EmIFVy
Agree with this. Our academic value system needs updating, and encourage scientists to take roles in public life / debates.
Of course doing research is the corner stone, but there are some aspects of academic rituals that could give way. https://t.co/n1avV9LNnr
@larsjuhljensen Yes. I consider funding part of the academic value system. But I think this is not the hardest part of the problem - there are quite a lot of funding programs that would like to see more public impact, outreach, translation etc. (Am familiar with EU, DE.)
@mikhailspivakov If/when mature & important enough, such software should be thought of as a research infrastructure, similar to a particle accelerator, a telescope or a genome database. It’s engineering for research, with a fuzzy boundary between professions. @BorisLenhard
@JennyBryan @bolkerb @JonStevensHall First also thought this was satire but at the same time it seems to be reality, see e.g. https://t.co/CiObbfplVp https://t.co/pn8pEtEDKs
Our study of ex vivo drug combination effects in chronic lymphocytic leukemia finds synergistic drug effects and genetic dependencies https://t.co/Tggo2hQEdd with @BrittaVelten, Marina Lukas and Thorsten Zenz
Join us for ‘BiocCheck-a-thon’, a week-long virtual hackathon to improve BiocCheck and the consistency and quality of Bioconductor packages! See
for details; starting May 18. https://t.co/cSSYV7nuAb
Use mathematical notation in your #RStats package’s manual pages with @wviechtb’s mathjaxr package https://t.co/xm7PaDwSPq https://t.co/A6M1nfZPKT https://t.co/BegwbrG8sN
A petition to the presidents of the European Council, Parliament and Commission to protect the European Research Council (ERC) in the EU budget https://t.co/crPK8t4ola
You can co-sign via https://t.co/I2RCExLrqk
@stephaniehicks @davisjmcc @_lazappi_ I agree that this terminology can be perceived to downplay postdocs’ expertise, energy and contribution; which would be wrong. OTOH I think it’s important to distinguish between postdoc and PhD-level staff positions. (1/2)
@stephaniehicks @davisjmcc @_lazappi_ Postdoc positions should come with more autonomy and research focus; staff with better pay & contract and more externally imposed responsibilities. (2/2)
@vallens @stephaniehicks @davisjmcc @_lazappi_ No, I mean people like software developers, database curators, those running core facilities or other scientific services. (Btw these are very important types of jobs that often have no clear trajectory in academia.)
@AdriAuladell @SherlockpHolmes But what’s the gas bottle for?
SAMHD1 recurrently mutated in mantle cell lymphoma, confers resistance to nucleoside analogue therapy in vitro. A useful building block in our understanding of a rarer cancer, found using ex-vivo drug testing. Great collaboration with T.Zenz @ViktorLu_1118 @marcobuehler et al.
@johnpoverington Are you using Excel 🤯
Postdoc position in computational biology, statistics, cancer biology: bring mass spectrometry & proteomics into cancer research and care! At @EMBL in Heidelberg. With many experimental and clinical collaboration partners. https://t.co/FGtTjR2u5W https://t.co/mZ014AyaEh
@DPereus @embl There are also PhD positions in these topics (and many others) via the @EMBL PhD programme https://t.co/qqViVhTCf2
@ewanbirney @embl EMBL is an international organisation with 27 member states and 6 sites: Barcelona, Cambridge, Grenoble, Hamburg, Heidelberg and Rome. Its mission includes research, service to the scientific community, technology & training https://t.co/qqViVhTCf2
@StephenEglen @nordholmen @HeidiBaya @alhufton I’m aware of this by @jimmy_wales: https://t.co/jLBx1AVObZ Using it in other parts of the world may be quite straightforward. It’s localized to English, main issue seems the TAN (transaction authentication number) you need from the testing lab to register a positive diagnosis. https://t.co/Mt2KkzLTbD
@nordholmen @betatim @StephenEglen @HeidiBaya @alhufton The app is not portrayed as a panacea, it’s one tool of many (masks, distancing) to reduce the probability of passing on the virus to others. A “high-risk” score in the app leads to advice to self-isolate and contact the health system (for testing, I presume).
The setting of @EMBL in Heidelberg is so scenic it even features on a completely work-unrelated bike tour during (much needed) vacation@home. https://t.co/30WnZl9aKH
This week would have been that of our summer course Statistical Data Analysis for Genome Scale Biology in the Alps (Brixen, IT). It had to be cancelled this year due to the pandemic - but we’ll be back in 2021!
Been thinking about this a bit. Its logical if you think of higher education as a private luxury and way to maintain a current social hierarchy. It’s madness if you think giving (your) higher education widely and diversely benefits everyone. https://t.co/F2A77UOm5N
@arjunrajlab @Chao_Jiang1 Good point. Thinking of -interactive plots -computational reproducibility through underlying code -easy updating & version tracking -many hyperlinks rather than just a few references Many scientists can do this on their own using e.g. RStudio, journal then degrades the product.
@mikhailspivakov @theosysbio @arjunrajlab @Chao_Jiang1 Like movie or book reviews….
@mikhailspivakov @CantoneIrene @theosysbio @arjunrajlab @Chao_Jiang1 There may also be a business model in getting readers (or their institutions) to pay for a service that highlights and perhaps predigests papers relevant to them. Btw people also pay for Google, Youtube, Twitter, albeit with their data & attention instead of cash.
@mikhailspivakov @CantoneIrene @theosysbio @arjunrajlab @Chao_Jiang1 Yes but there the model is to get those who do all the hard work do it without pay or other good enough incentives
@Sci_j_my @AcademicChatter On nepotism in academia: “…[the data] make a pure meritocracy seem implausible, suggesting the influence of nonmeritocratic factors.” So, subnetworks (disciplines, regions) with more meritocracy should be more competitive. (also: arbitrage opportunities for clever recruiters)
@Sci_j_my @AcademicChatter It may be very hard to get completely rid of such biases (https://t.co/uSDImGyLSB), but for those not so lucky to do their PhD in a high-prestige place, there are postdocs, internships, open contribution consortia (e.g. @Bioconductor, #RStats) etc. to make yourself more visible
@LH33837868 Congratulations from me, in this way, too! Happy to see you take this step, in such a productive and exciting environment. Wishing you the very best!
@zolotarg … and by @SherlockpHolmes, who also picked the roulette wheels and the little demons.
And, yes, the book is evolving in time, the online version is continually being updated https://t.co/obiejJIy4R https://t.co/Qjlp7WmCf0
@mikelove @mikelove’s visit in Heidelberg was 17 Sep 2012 - 28 Feb 2013: the birth of DESeq2, amid lots of animated discussions also with @s_anders_m, including some of them in the cozy pubs around town.
@researchremix @lgatt0 John Claerbout said it in 1994 https://t.co/eb5zOn78CM
@AedinCulhane @slavov_n @chrashwood The caterpillar and the butterfly - same genome, different expression of it. https://t.co/6v7Vg93QQZ
Congratulations to Robert! Exciting. https://t.co/bSHIxFn1Oc
This attitude is too prevalent in the life sciences. And detrimental to scientific progress and overall impact of the field. It encourages scientists to do lots of little separate projects and papers rather than pooling skills and efforts to do something big. https://t.co/trJmDUa7JW
Pubmed is starting to index preprints. This is currently only for NIH-funded research - but definitely a step in the right direction. https://t.co/lggxDSB01k
Is anyone aware of how multiple (remote) people can interact with the same R Studio Server (Pro) session? Everyone types into the same terminal and everyone sees the same outputs? (I.e., kind of hybrid between zoom screen sharing and googledocs, sorry for brand name dropping)
@PavelTomancak Heads of state or government (Council) did this (in particular self-styled frugals and nationalists), not “the EU”. There is still hope that this will look different after parliament and commission are done with this.
“Up to 30 additional Alexander von Humboldt Professorships in the field of Artificial Intelligence are to be filled in the years up to 2024. Award funds €5 million in experimental disciplines and €3.5 million in theoretical disciplines for five years.” https://t.co/9T74IyrMY7
@PavelTomancak @ONeyrolles Personally my mileage has been different, FP6, 7 & H2020 have funded (software) infrastructure and engineering work that would have been more difficult to do using other sources; and I met people across the continent who have become collaborators and friends.
@ONeyrolles @PavelTomancak There’s a little bit more to it than that: https://t.co/C3B87vyUjc (and yes of course yesterday’s budget compromise by the council is upsetting and flawed)
For similar reasons, software (engineering) also needs to be taken more serious in academia and research. https://t.co/QGTrvresBP
@lawrennd Good point, Neil.
I was thinking here more specifically of scientific software and software as a research output, when reading the subtitle “As software eats the world, more […] are being nibbled at by their computer systems”
Looking for summer reading? This little book on tactics people use in public discourse https://t.co/b8WZPDFZgn was written 200 years ago, and just as relevant in times of Twitter & FB. Short, fun & mildly useful (I recommend the edition by @acgrayling, which is a bit hard to get)
Great opportunity in an ambitious and high-powered new institute in one of the best cities in the world. https://t.co/MJ4SqklCZu
Gaussian process fitting with Gamma-Poisson (NB) likelihood with applications to single-cell RNA-seq data. Sounder inference without hacky log(n+c) transformations. https://t.co/WOFNXnIP5p
This little guy has one of the best daily commutes to kindergarten @EMBL (which of course operates under strict rules for the ‘conditions of the pandemic’) https://t.co/4GqzNr0ml3
Do you answer emails FIFO or LIFO?
I’ve now seen a few scientific conferences that were basically series of zoom meetings & webinars, and quite effective. I wonder how things will be a few years from now. 3D immersive virtual reality with avatars etc.? Imagining scenarios for some of my favorite meeting series…
Intuitive explanation why|how models with very many parameters (e.g. deep learning) can be useful https://t.co/yX6gzbxQoR
@lgatt0 I used Googledocs to read the PDF and then the comment function. This was for a signed (non-anonymous) review. I got the impression it was more useful for the authors than a traditional report, easier and more fun for me, but the editor did not know what to with it.
Or many other committees I will not name…. https://t.co/6lMuY57q15
By @const_ae https://t.co/CHhiPrdwd1
@sabahzero I stumbled into a postdoc in cancer genomics as a physicist since it was novel then & fun. I expected going to industry eventually, was skeptic about having ideas or doing glamour science for academia. When I saw how people needed statistics & software, I applied for PI jobs. 1/2
@sabahzero I’m grateful to many colleagues for being lucky to met them in the @Bioconductor project & beyond, for being fantastic role models, and for teaching me what science was really about. 2/2
@sabahzero Btw- I am very happy with that choice, but am sure there’d also have been different, but equally exciting and rewarding paths in the private sector.
The movie analogy for writing papers. - Introduce all relevant characters and plot lines early on - Get your audience excited to want to read the paper early on, and do not lose them in the middle - Don’t arbitrarily rename characters mid-plot, or give them no name at all https://t.co/gDcG8wWnOo
@KLdivergence @igorpianist Pianist, tweets on classical music performed by him and others
@ChenxinLi2 Yes! There’s also a little discussion of this, and bar, box, violin, beeswarm, density and ecdf plots here: https://t.co/Oy0SAFxUJm (w/ @SherlockpHolmes) https://t.co/v2ddKUXHPj
A tweetorial on the t-test: I often hear people being nervous about normality assumptions, avoiding using the t-test because they think their data aren’t normal, or even running tests “against normality” to show they can’t use it. There is rarely a need for such worries. 1/5
The t-test works just fine for data quite far from normality. There is however, another assumption that is far more critical: independence. If there are correlations, or batch effects, then the results of the t-test (and pretty much any basic test you can think of) are unreliable
Applied to correlated data, the p-values from the t-test will be all over the place. Often, too small, leading to false discoveries. I put up a little shiny app https://t.co/H56rHYt1J6 to demonstrate this. 3/5
Even for very long-tailed distributions, the test’s calibration is fine. If anything, the t-test gets more conservative. If there are correlations, however, there is a strong effect: an abundance of (spurious) small p-values. 4/5
This means that you can use the t-test for quite non-normal data; not that you should. That depends on whether the mean is a good summary. For very skewed data (say, income distributions in a country), shifts in the average may just not be the right thing to look for. 5/5
@tamas_schauer This is hard. Need to carefully decide on your hypotheses and what you want to show, tailor your statistic (say, some “score”) and use resampling. MW U-test is most likely not the answer.
@MKrzywinski @naturemethods The Points of Significance column has been a great way to give accessible, short overviews over practical statistics topics https://t.co/lCd3meriDf What are your plans for its future?
@MarioNiepel There’s a little bit about this in https://t.co/TqSJKI8xPs esp. Sec. 13.4.5, but the main point is that the ‘replicates’ need to cover the population that you want to make a statement on. If you’re happy to make a claim about one particular well in one particular experiment (1/2)
@MarioNiepel then those 1000s of cells in it are your replicates. But if you want to make a scientific statement about this kind of cells under that kind of treatment in general, then you need to replicate across multiple samples from those cells, multiple repeats of the same treatment, etc.
@pmelsted Et voilà: https://t.co/Re8mkAy2qo Incl. DESeq2, IHW and many other preciosities.
@pmelsted I love the one for #Eurobioc2020 (by @jo_rainer and @drisso) hosted by Univ. Padova, showing Galilei, one of the leaders of the transition from the dark ages to a more hopeful and rational era, using telescopy, and founding modern science. https://t.co/Q8QOOXZIdc
Scientific writing: do not use synonyms for the same concept. It may be clear to you that these different terms all refer to the same thing, but readers will start wondering whether there are (subtle) differences, and as a result misunderstand, be frustrated, loose interest, …
@markowetzlab Why, you could just write ‘move the boundary between the known and the unknown in a relevant place by a noteworthy amount, using appropriate methods and a reasonable approach’.
@jjtokyo Not sure it’s such a matter of language, culture, or everyday writing. I think it’s like with the plot of a movie or book with many characters (think War and Piece, Game of Thrones): you don’t want their names keep changing all the time.
“Gender Roles and their Impact in Academia” EMBO | EMBL | HHMI conference (online, 13-15 Oct 2020) Abstract submission until 14 Sep 2020 Registration until 22 Sep 2020 https://t.co/bmzc9Qgatf
@MariaHondele Congratulations, Maria! A great recognition and opportunity.
Thank you for the kind words, Rahul. Glad to see it useful! https://t.co/KIcCvDn8gv
An important building block for limiting the spread of the virus, the Corona-Warn-App in Germany with 17.8M downloads so far; country has 83M inhabitants. It also automatically displays your test results (I got a negative one recently). https://t.co/yQWD5KxIdU
Logical reasoning with uncertainty https://t.co/RGQ1f6FFLj
In this thread: my subjective mini-review and recommendation of “Working in Public: The Making and Maintenance of Open Source Software” by Nadia Eghbal (https://t.co/itRnNSqJur). Don’t expect … 1/4
… ground-breaking revelations or definitive solutions. It’s an attempt to systematize and put into words many observations that may be obvious to old hands; it provides interesting anecdotes, some useful statistics and a few pointers at how people try to move ahead. 2/4
Some bullet points: -a classification of project types: federations, clubs, toys and stadiums -requirements of maintenance & support -code is alive and in constant need of maintenance; there are associated hidden costs -the community- and behavior-changing impact of GitHub 3/4
-assessing the health of a project -managing over-participation, dealing with too much noise, with extractive (negative net impact) contributions -community & contributor growth vs “the myth of the person month” -developer attention is a scarce resource. 4/4
@BoulesteixLaure @MaartenvSmeden You could try simulations? https://t.co/doxg1VlIcP Also, t- and Wilcoxon-test have different null hypotheses, and are differentially sensitive to different alternatives, so they’re not really plug-in replacements.
Live session by Friends of the ERC on Thursday 24 Sept 9.45 - 10.30 https://t.co/abIa0O7ULg with speakers incl. the president of the ERC, J-P. Bourguignon, et al.
Idea: every scientist has their zoom talks online and a GitHub-like issue page for questions. Organizing a conference = creating a playlist
@GaloGoig @ZaminIqbal https://t.co/7cakqf4YIs
@grimbough Interesting. Mine are either classics (Vivaldi, Bach, Mozart, Beethoven) or current EDM.
Congratulations to a great scientific collaborator, friend and colleague (https://t.co/4mkGBNH6bQ) https://t.co/HxYJB3oHAM
Submit a talk, workshop or poster for the European @Bioconductor Meeting 2020 (14-18 December, online): https://t.co/umyxIiqRCC https://t.co/4s8WloG6Qh
The joy of a day uninterrupted by meetings or emails and dedicated to writing or revising a manuscript.
Would you down-rank a project proposal if they use the term AI where they could have said machine learning?
“In engineering, a published paper is an advertisement of scholarship but the electronic document can be the scholarship itself. Forty years ago … paper documents were adequate. No more.” Claerbout (1994) https://t.co/fiDKWbQ3wh https://t.co/hEPJYKaLT3
@drisso1893 As many in the replies pointed out, the issue really is about claims of “(some opportunistically available data) => (unspecified black box method) => (wonderful result)” and whether the choice of words correlates with this argumentation pattern.
Interesting comment by Esther Duflo on the need for better multiple testing methods https://t.co/XtmxSC1bZ2 (31:44-35:00 min) Current approaches are brute force and waste data collected with great effort. Need to use the data as efficiently as possible.
Bought an Audiobook so the 4-year old could listen to it on his single purpose MP3 player device. What I got from Amazon was an AAX file (Audible), which can only be played on proprietary apps. As is, useless. Here’s a way to convert into a series of MP3 files: (1/3)
First, follow https://t.co/32suLOTn81 to convert into a regular M4B file (a container format). 2/3
Second, use m4b-tool by @sandreas https://t.co/3QO5rU1cXx to split into individual MP3 files. For me, the docker version worked, and I renamed the file to something simple like a.m4b to avoid apparently fragile filename string handling. Junior is happy. (3/3)
Correction: the twitter handle of author of the m4b-tool is @spiessa and the point is to split into one MP3 per chapter for easy chapter navigation.
@SherlockpHolmes @dror_barak @knstnr @CFlensburg Or you can do:
(in one line; to circumvent Twitter’s URL shortening, I post this as an image…) https://t.co/t4Dq00FCoc
Such a happy twitter timeline this weekend.
Here’s an R script that does some image contrast transformations to make the hidden message more visible https://t.co/DJnSV707qp (very simplistic - probably could be improved by more sophisticated computations) https://t.co/0AIiiB0evy https://t.co/N6edtU7fPa
Congratulations, Piero - Looking forward to seeing growing this into a genomics powerhouse! https://t.co/LkUxgBdTGo
@JShendure @minouye271 At a multi-candidate symposium for a professorship at a prestigious university, I gave a completely overloaded talk. Host had to interrupt me after I ran over time. Then had to do 1:1s, meals etc for two more days keeping a straight face while obviously being the hopeless one.
Two exciting postdoctoral research opportunities in machine learning in cancer research https://t.co/m9yRXyMtT3 https://t.co/UzL3z4zDCW
Here is the right link: https://t.co/rnUvu7R1wE https://t.co/ZYyBnWS0UV
The full link is https://t.co/rnUvu7R1wE Job-ID HD01864
@florianjug A central issue for OS developers. @nayafia has written a more extensive (and more politely worded) study on this (incl. data & lots of examples): https://t.co/2dRxmeEg8k - worth reading.
If you’re wondering about the difference between the reported 94.5% of the Moderna vaccine and the 90% of the Biontech-Pfizer: fisher.test(cbind(c(90,5), c(84,10))): p-value = 0.19 95% confidence interval: 0.63-8.30 i.e., too early too tell (1/2)
Moderna reported 90 placebos among 95 cases, B/P 84 or 85 (90%) among 94 cases. fisher.test is the R function for https://t.co/6N3BYQ1VAH (2/2)
@jcbarret Thank you Jeff, for pointing this out. Silly mistake! Got too excited about it. I deleted the tweet with the wrong counts and replaced it by https://t.co/8iwyE6bNW5 The conclusion is the same.
@RuneLinding Thanks, yes, this is very encouraging. Also the easier storage.
@JBPingault @ewanbirney The Fisher test result implies that confidence intervals (CI) are widely overlapping and based on the current information, the two treatments have indistinguishable efficacy. It’s possible to compute CIs but this involves subtleties/choices that I considered needless here.
@santa_care @ewanbirney True if you rely on GP offices etc for delivery. But to get the needed throughput, some places (e.g. Germany) are planning big vaccination centres in exhibition halls and the like, where having -80C freezers should not be the bottleneck.
A run in the woods around @EMBL after a day of zoom meetings https://t.co/A0MYGWbQUI
@nicolesoranzo @embl Of course! Here’s the full resolution file: https://t.co/rnRTFNtYYb
⬇️ Position as a research infrastructure scientist in my group available to work on data analysis workflows and user experience on the German Human Genome Archive (GHGA, https://t.co/uesXwiZw21), a national scale archive for human genetics biomedical research. https://t.co/9IsMZAAisB
@watermicrobe …. amazing & pivotal mentors of my early career include Julia Rice at IBM, Annemarie Poustka at DKFZ and Janet Thornton at EBI -
and several other ‘senior’ female collaborators that have been crucial to my work.
Long video calls are tough but Skype for Business is tougher.
Should have dropped every second word in the tweet to make it more authentic. https://t.co/0nVx0Dg4k0
@arjunrajlab @robinsall To be fair, the pseudo-objective, pseudo-normalized quantitative nature of this metric is an invitation to warp it.
@NateSilver538 Not claiming particular expertise, but simple algebra derives the following “reverse engineered” numbers from the AZD1222 press release, which suggests that the subgroup difference is real (https://t.co/I85bMUgIj7) https://t.co/o3POsxZd1k
@NateSilver538 The point estimate of 90% based on the numbers 35, 39 has a large confidence range, but the fraction does appear different from that in the FF group.
@JamesWard73 @ewanbirney @alexselby1770 @jamesward73, do you refer to https://t.co/FxdRUwwdBB ? @ewanbirney refers to https://t.co/RHE7TZ4F3J Difference is the definition of efficacy. I used what BioNTech/Pfizer et al used: controls/(treated+controls), which is between 0 and 1. The others used (control-treated)/control
@JamesWard73 @ewanbirney @alexselby1770 @MKing7403 Thank you for digging this out! Now the incidence rate ratio IRR is a complex beast that also depends on the total number of study participants and their times at risk, not just those who got infected, so unless these numbers are known (or cancel out by symmetry), … (1/2)
@JamesWard73 @ewanbirney @alexselby1770 @MKing7403 the ‘reverse engineering’ problem is underdetermined. 1-IRR is not equal to either of our simplistic definitions, although @alexselby1770’s is closer.
FWIW, for Pfizer/BioNTech data (tr:8, ctrl:162) the values for the two definitions numerically coincide: 162/1700.95154/162.
@NateSilver538 Correction: the numbers in the HF regimen are even smaller than I estimated above. A recent media report (https://t.co/HuMGYfdcZ5) indicates 3 infected cases in the HF group, and together with a better approximation of the efficacy estimator, 1-treated/control … (1/2)
@NateSilver538 … (@alexselby1770, https://t.co/VEOlmoPMeB), this leads to below contingency table. Similar to what others said, this indicates that until there is more data (better,bigger study), we can’t be certain that the claimed higher efficacy of the HF regimen isn’t a statistical fluke https://t.co/jIjVXVGiAD
View from Koenigsstuhl to downtown Heidelberg, where it’s foggy, dark and cold. The bike ride up here almost felt like an allegory of 2020/21 (I hope) https://t.co/AaCZbPk5If
Register until Monday 7 Dec for the Bioconductor Europe meeting (14-18 Dec, online). Exciting lineup of invited and contributed talks, workshops, posters and BoF https://t.co/KF45WhIVVK https://t.co/7JMGWOQhUt
Does anyone know how to permanently get rid of ‘suggestd’ on MacOS BigSur? This built-in spyware by Apple regularly clogs up most of CPU capacity on my computer. Even (force) quit only makes it go away for a short while, then it reappears to make every other app sluggish. https://t.co/UMtD8kNzhh
@stephaniehicks @mikelove @jhubiostat #OneYearAgoToday, European Bioconductor Meeting 2019 in Brussels, the picture is from a wonderful conference dinner at Hof ter Musschen. https://t.co/5WFGLqTGuW
@stephaniehicks @mikelove @jhubiostat Local organisers @lgatt0, Axelle Loriot, @lievenClement . In the foreground, keynote speaker @andreamrau, and Simone Bell.
Beethoven 250 years anniversary concert 2020-12-17 by Daniel Barenboim & West-Eastern Divan Orchestra: 3rd Piano Concerto and “Symphony of Fate” No. 5, presented under Corona conditions, as a symbol of hope: https://t.co/trUL2QBhql video: https://t.co/f54mKkQ1jT from 13’40”
@cshperspectives Here’s a background story on @cshperspectives’ statement from The Guardian. The current system has been deliberately devised by entrepreneurs to extract money from public research funding, and scientific communities have been docile enough to play along https://t.co/8bGGvOkZE3
@StevenSalzberg1 Time & more data will tell, but for the record, the panic was set off by the UK government on 2020-12-19 trying to find an excuse for a belated policy U-turn: https://t.co/eYuRjrrHRw
@drisso1893 Hmm, this (no direct pay for submitting an article) is in my view a good aspect of the system. You are paid with a tax-funded salary and indirectly with career prospects, grants etc. Anyway, the real cost of producing a manuscript would be hard to account for, but certainly high.
Why is it important that method papers are accompanied by functioning software? ‘Conceptual’ methods papers without software are analogous to patents without intention to make a product: they make wide-reaching, vague claims in order to occupy a certain intellectual space…(1/2)
…disincentivize others from working on it, thus slow down further development, and provide no benefit to society. Software implementation forces authors to be specific about every single choice, and provides a benchmark for others to compete against. (2/2)
@SeriousSwann @drob Thats a valid question, but in my view not that different from the same question for the quality of the writing, figures etc of the research article itself. This can also be more or less elaborate, some people put in an enormous effort, others just do a minimum.
@GarfieldLab Sometimes a single publication really moves science forward, while series of others are inconsequential. Good employers recognize that. So I think rewards are there.
Also, I think most scientists are in it to do the right thing, not just to have comfortable but pointless jobs.
@HenriquesLab Year in Edinburgh 1990/91 studying physics & meteorology. Eye-opener and first bigger view of the world and int’l mobility for this working-class boy from a farming village in rural Germany. Kept strong ties to the UK and went to Cambridge in 2004 for my first PI job (EMBL-EBI).
@HenriquesLab Took up Scottish country dancing, white-water rafting, mountain biking and took a general relativity class from Peter Higgs.
Thread (grim): “B.1.1.7 was repeatedly capable to proliferate under lockdown measures sufficient to suppress other SARS-CoV-2 lineages.” “Control of B.1.1.7 will require stricter measures than applied during the November lockdown (in England)” https://t.co/unxU3v6wmQ
@robertbrussell It seems to be both “normal virus evolution” and scary for humans: https://t.co/unxU3v6wmQ and https://t.co/jypkphDGUI
For my local area followers, here’s a link to a rapid-test centre sponsored by the city of Heidelberg: - drive-thru - result in 20min - antigen-based, claimed sensitivity 98% - near Bauhaus in Eppelheimer Strasse https://t.co/7KCIOtctwe
Re branding/marketing of new statistical ideas and how computer scientists always have the cooler names… how would they call post selection inference?
@daniela_witten Just Monday morning? Aren’t academics’ inboxes like a hydra that grows two new emails for every email answered?
Cycling around @EMBL Heidelberg January 2021. Above the ice zone, the snowy paths are quite pleasant. https://t.co/pbXihE7g7L
Deadline 31 Jan: ARISE fellowship program for professionals with background in STEM who wish to advance technology development in the life science research infrastructures. https://t.co/0Bnh8eYESe For my group, we are looking for an engineer who can improve the usability… 1/3
of the new GHGA database (German Human Genome-Phenome Archive https://t.co/uesXwiZw21) and the usefulness of its data, by constructing (e.g., R-based) workflows for data analysis, teaching them to prospective users (biomedical researchers) and … 2/3
working with the GHGA core developers on an efficient API.
3/3.
Reading the EMA’s Public Assessment Report on the BioNTech/Pfizer vaccine cominarty feels a bit like reading a PhD thesis, although with bigger scope and immediate impact https://t.co/fspsxWoqgo https://t.co/SaPM7F3uHq https://t.co/vCfbHjxrjm
TFW when the page proofs of your carefully LaTeX-typeset maths manuscript come back from the commercial publisher full of ugly spaced and font sized formulae and content-changing “journal style” edits https://t.co/jnGjxv7eHk
(of course 48h deadline after 3-years review process)
It gets more hilarious. After mutilating our beautiful equations into their corporate typesetting program, they expect authors to work with their awkward substandard web-GUI thingy to fix the mess for them. https://t.co/AhGHxZD2z8
Now the academic review process was excellent and very helpful - am just rather unimpressed with the production editing.
👇 (I can’t judge all the details in these analyses, but in any case it is an example of the need for noise modelling & deconvolution from measured values to the latent variable of interest.) https://t.co/Gbvw5dRdGN
The Benjamini-Hochberg algorithm is so engrained in bioinformaticians’ minds that many think there is a natural 1:1 correspondence between ‘raw’ and adjusted p-value.
In fact, … (1/2)
… the ‘raw’ p-value is a simple per-test property, independent of how many other tests you did and what their outcome was—whereas an adjusted p-value depends on all that, as well as choice of multiple testing method.
Tip: do not use a boxplot to visualize data with a bimodal distribution.
More on this: https://t.co/ONsCcRaWrD
It’s fun to lighten up your writing tasks with some programming. That’s one reason why I like LaTeX. Here comes the next level. https://t.co/nG92MWePAo
@ewanbirney @NateSilver538 Why would the clinical trial be ethically problematic if the real world rollout is not?
@Francesco_i0ri0 @overleaf I don’t - settled on Googledocs for that… (does anyone know an alternative?)
I do fondly remember using subversion and LaTeX for collaborative EU network grant writing with @lawrennd and @MagnusRattra once in 2011… https://t.co/ErfnCAX0gR
@OliverStegle @Francesco_i0ri0 @overleaf LaTeX and github/lab for the supplement rocks. Anyway the supplement is where all the real action is.
It’s amazing how far you can get in scientific writing by always being clear about sufficient versus necessary conditions. I.e., does A ⇒ B, or B ⇒ A?
@theosysbio What are your best examples for the usefulness of these concepts in biology?
@theosysbio I’ll start with the Voronoi-Based Segmentation of Cells on Image Manifolds in CellProfiler (https://t.co/fNlTMJ9Ssh ).
And of course all the embedding methods: t-SNE, UMAP, phate, sleepwalk (https://t.co/9fXoAi59PU)
@BioMickWatson @luispedrocoelho @bioinformer @Microbes4ever Yes… if the data do not come in physical units (such as mol/L) but just as counts, there needs to be some anchor or scaffold to normalize across samples, such as most genes, total sum, or spike-ins,… Otherwise its hard to see how to do any quantitative analysis.
@BioMickWatson @mikelove @luispedrocoelho @bioinformer @Microbes4ever Assumptions can be sufficient or necessary, it’s useful to check how bad (or not) it is for the final outcome if they are slightly off. One assumption that is often underappreciated is independence - absence of which means “batch effects”.
@mikelove @klmr They’re slowly starting to address this… see e.g. https://t.co/YdXsF1ONSb and https://t.co/PmMzT5iGdV
@notSoJunkDNA @NEJM @embl Disgusting. Thank you, Nicolas, for pointing this out. We will see what can be done about this. It is against EMBL rules and regulations. On the journal side, I expect there’s a process for correction. There may also be legal implications.
Snowy winter’s morning at @EMBL Heidelberg, at a crisp -10°C https://t.co/JEt36DzmGj
I got a proper microphone for recording my online lectures rather than using the airpods, and it is a gamechanger. https://t.co/AwR21u70q7
@mycroscopy The main thing to share is the book, https://t.co/VeF6EmIFVy (print) https://t.co/obiejJIy4R (free online). There are also slides and recordings, e.g., https://t.co/pPsljE6iFm (2021, emerging) https://t.co/PqjwN2ICBq (2020, with @SherlockpHolmes, whose recordings are recommended)
@fabian_theis Have fun and enjoy! It is a precious time.
@BioMickWatson 2030s: realtime analysis of a zettabyte/s planetary datastream of a billion nucleotide sequencing proteomics imaging cloud drones biosampling scorching megacities, country-sized agroindustry fields and a few remaining wildlands
Great opportunities to start your lab also for researchers in computational biology, bioinformatics, biomedical machine learning https://t.co/Ex7Jy6Nb7N https://t.co/n9Lc5XK5lI
If your analysis of high-throughput data (RNA-Seq, proteomics, gRNA screen) yields non-redundant values for effect size and statistical significance, do not squash them into a single score and look for the best selection cutoff. Define your decision boundary in the 2D space.
@extended_gene https://t.co/duVQS6j59Y
Calendar invites should also contain the videocall link/identifier and the agenda.
Sending such information in separate emails (and even at separate times from different senders) is unfriendly.
Am trying to get an overview over methods for dimension reduction or feature selection that do not just go for large variation, but for large variant consistent between available replicates. Any pointers welcome!
1st reply is of course the excellent https://t.co/WTKopdZWlp project
@stephaniehicks @anshulkundaje We need more appreciation of engineering in science, as projects get bigger and more complex. Visualization and UI are examples, but there is also data management, professionalization of software etc.
Originality & individualism are important, but overemphasis stalls progress.
@stephaniehicks @anshulkundaje … and there are good moves in that direction, e.g., the ARISE program by the EC and @EMBL https://t.co/CVgkLRupI1
@embl @emblebi “A scientist has about 50 years to do their research; taking a short time out … to be at home for whatever reason should certainly be possible.” A very wise and reasoned demand. We’ve still some ways to go to normalize this. The hamster wheel of grant funding, … (1/2)
@embl @emblebi productivity reviews and multitasking supervision of (thesis) projects seems very much woven into the current system. (2/2)
@LeviWaldron1 Time for email bankruptcy? https://t.co/y74nLerfsX
I took a dive into the subculture of (good) toddlers’ bikes and it’s amazing. The most favourite brands are sold out and not even taking orders, everyone has long delivery times, and used bikes are traded on ebay at prices above the catalog prices, and are gone in minutes.
@theosysbio Yes. Although the trendy brand here is Woom (from Austria). Both equally out of stock. I got a used Woom3 for the bigger boy via ebay today, from someone 60km away. Littler one gets his Woom2. Agree training wheels are counterproductive, better go straight from balance to pedals.
Fantastic opportunity @EMBL Cambridge https://t.co/BfWqwx0Pfm
This looks like an exciting opportunity in biological data science at Cellzome, a company where I and others at @EMBL have several great collaborators (and not far from our campus) https://t.co/Xdqwhhjgxj
To get stuff done, any administrative organization needs good processes and good software, and it seems few managers understand both.
@arjunrajlab Perhaps a more meaningful dichotomy is between observational study, providing correlations/associations, and interventional experiment, directly providing causal relationships?
@arjunrajlab (and of course as with many dichotomies, this is a caricature; observations can be used to exclude certain causal relationship hypotheses, or together with prior knowledge to choose between several models.)
Call for Abstracts useR! 2021
deadline: 15 March https://t.co/InExeOPclf https://t.co/UQVvS5inVY
@larsjuhljensen @MicrobiomDigest Given the amount of time (=money) behind any manuscript, it ought to be economic, for the overall science enterprise, if someone spends a few more hours on feedback. (1/2)
@larsjuhljensen @MicrobiomDigest OTOH, reviewing is a sparse resource, and it makes sense to direct it to where it has most impact. My vote: scrap peer-review as gatekeeper, preprint everything, post-publication peer review (probably partly professionalized), overlay journals. (2/2)
@drisso1893 It’s identity politics in science. And not only with journals, also PhDs, faculty recruitment, grant panels. Some people seem to need it. Certain leaders use it to assert their power over a discipline. But I am optimistic: scientific progress comes from recombination of ideas.
Welcome Moritz to Heidelberg! https://t.co/9R3uBXdmCH
Machine learning (supervised) reminder: the cross-validation (CV) for parameter tuning is no substitute for CV to assess classification performance. Need to do two nested CV loops then. https://t.co/POz9GbFPy0 https://t.co/y0l5LefO9u
@markowetzlab Wow! Well spotted. So deep from the mists of history.
https://t.co/tIzsLcmjAM is a link without paywall
@markowetzlab These authors didn’t mince their words. From p.20: https://t.co/UGDYtzOyFo
This. True also for much of statistics and data science more generally. https://t.co/qo4SJUDwZq
@JD_Buenrostro The High-Dimensional Data Analysis section in https://t.co/cpANgexQSL
Senior Job in Science Communication @EMBL: https://t.co/BDNFEfynlj
When did spending eight hours on zoom become a “retreat”.
@ewanbirney It’s worth it. It also helps clearer thinking about higher level design and messaging issues with your graphics. And works very well together with dplyr for data manipulation (which is a big part of doing visualization).
I tried to give an intro here: https://t.co/CYOoCY9X9D
@tpoi To be fair, science today is more collaborative, with bigger projects, more infrastructure, more need for training & self-administration. The idea of a lone genius toilling away in a chamber is romantic. The right trade-off between talking and doing remains hard to find.
Social media https://t.co/mCTb2HLX5A
Functional Precision Medicine in Blood Cancer Symposium (virtual): 25 and 26 March 2021, 12:00-19:30 CET (7:00-14:30 NYC) https://t.co/OtHJSNrqd2 https://t.co/Lgkd9NlwXg
I am one of many deeply worried about the further delay in Europe’s vaccination program caused by the AZD1222 inquiry. This fact sheet by the German regulator is helpful. The population of 20-50yr olds has not yet been highly represented in the big AZD1222 rollout in the UK.(1/2) https://t.co/AR3K61iylT
Ultimately the assessment of this situation seems to boil down to a choice between utilitarianism and the precautionary principle, a tricky political and PR task. (2/2)
@d_spiegel “The principle has instead moved countries away from one risk (blood clots) towards another (lower vaccine coverage)”. This is complex. The people mostly affected by one risk (older men) are different from those of the other (younger women)….
Recording of the 2021-03-18 Bioconductor Developers’ Forum on a new OO system for R (as proposed here: https://t.co/rjL6mvIwpa) with @grimbough @lawremi @hadleywickham @jimhester_ R.Gentleman @MMaechler @henrikbengtsson
@Y_Gilad Do a PCA analysis instead.
Am noting in collaborators’ writing the idea that introducing lots of abbreviations makes your text look more scientific. To me, it just revolts readers.
@pedrobeltrao @larsjuhljensen @OliverStegle Faculty are 25x more likely to have academic parents… - how much of this is voluntary? - how much boils down to grooming, networking, nepotism? - could academia do better if it were more inclusive?
Feels personal to me, as 1st generation, & offspring of farmers and workers.
@larsjuhljensen @pedrobeltrao @OliverStegle First to graduate from high school 🙂 (abitur, baccalauréat). I’ve not cared much either. It’s probably had good and bad consequences, as many things do. Both you and I are lucky to have grown up in countries with relatively egalitarian education systems.
@IanSudbery Maybe it’s often not so much about space or flow, but wanting to be a seen as having discovered a new phenomenon, created a new method, invented a new field, … which of course need their own new name (=acronym).
Upbeat piece on RNA biology in the Economist. Coming out of the shadows of protein and DNA 😎 https://t.co/eOlz8mckKi
@BodoBrueckner Sneaky use of the passive voice.
anima sana in corpore sano https://t.co/6r8MBRLer4
. @mikelove on the origins of DESeq2 in the winter of 2012/13. Most of the time, Heidelberg is of course a bright and sunny place. https://t.co/Hxi7NcafpV https://t.co/nmS9IJBEjX
@mike_schatz @mason_lab Of course, https://t.co/DR6nqAQdV6
@ClaireJStandley Hi Claire, what’s the source for this, and in general for statistics on vaccinations, vaccine stocks, and deliveries?
@ClaireJStandley @ECDC_EU Thank you! There’s also https://t.co/L6TS91fVIP (by the federal govt), it seems to imply similar numbers. Is this a problem of holding back intentionally for people’s second doses (unreasonably…), or deeper problems of bureaucracy or skepticism?
@ClaireJStandley @ECDC_EU The “Impf-Fortschritt” graphic on https://t.co/L6TS91fVIP claims that vaccinations went ahead at same rate last week as all of March. Rates are expected to rise in April as more doses are delivered (which mostly hinges on BioNTech). https://t.co/fpyKLZqLrf
@AlexGuseman This can work esp. if staying in the same field. When I moved from industry to academia, and from cheminformatics to cancer genomics, a formal postdoc application worked well for me. Also, in the meanwhile I recruited several great field switchers via the formal process.
@AlexGuseman “Informal” seems to benefit people who are already well-networked, a more formal recruitment to be better for diversity, openness and transparency.
6 days later - icy winds from the North, apparently more to @mikelove ’s liking. https://t.co/X7harFTJEA
What’s the best place to disseminate Rmarkdown | Jupyter | … notebooks reproducing the analyses of a paper? - Paper’s supplemental material - GitHub - CodeOcean (e.g. DOI 10.1038/s41587-019-0136-9) - Bioconductor - …? There seem to be pros/cons for each.
For the record, I think it’s useful to distinguish between two objectives: - reusability (of a tool, method) - reproducibility (of a scientific claim, discovery) For the former, there is @Bioconductor & Co. My above question is about the latter.
@grimbough Thanks, Mike! Although I think that for many of the papers there, the software ideally would not only be reproducible (“one-off”) but also reusable, which would speak for putting it into a package repository sensu https://t.co/OSv5utUvfP
Vaccination rate in Germany is picking up, as deliveries are finally coming in. Double the number yesterday than any day before: 656,357, that’s ~0.79/100 population. Should soon be similar across EU? https://t.co/wiGCJArX9v https://t.co/Z6PDf8O8Bc https://t.co/9BndwxYWLt
@effigies Brilliant. I’ve a similar text as an email shortcut. There are many reasons for not getting drawn into 1:1 support via email and insisting on doing it via forums. Many of them also outlined here: https://t.co/9JxzCbgTOQ https://t.co/VTnYFO8Byf
Don’t be that kind of academic. https://t.co/fFfrCFs0fE
TFW when the author/LaTeX typeset manuscript on @arXiV looks way better than the professionally edited version in the fancy for-profit journal.
Methods. We processed samples 1-49 using (software) V14.4, samples 50-89 using (software) V15.5, samples 90-128 using (software) V14.5.
@MagnusRattray Absolutely. Just wondering whether as factor, ordered factor, real number, or as picture for a deep learning net.
Full Professorship in Molecular Biology with a focus on “Engineering of Molecular Systems”, in sunny Heidelberg https://t.co/OI1GHs0HPP
@StephenEglen @lgatt0 @nordholmen For beginners, I’d do tidyverse (incl. dplyr, ggplot2, readr) for data I/O, transformation and graphics, and skip the base analogs (predecessors?). Broadly along the lines of https://t.co/CuXjBdxSw0 tibble and data.frame are largely exchangeable, the former is more consistent.
@StephenEglen @lgatt0 @nordholmen For numeric matrix like data, as in omics, @Bioconductor’s SummarizedExperiment is an importanr concept. There’s a brief discussion in https://t.co/61i6IdCMks and Sec.13.11
@iscb_scs @embl @iscb @iscbsc @GonzaParra_ @CuypersWim @cuypersb @CleidyOsorio @VictorGrentz @caicebalt @KhalGabbo @Mina_2912 @krmsalazar @pradeeperanti Hi, thanks! Indeed I got a PhD on an attempt to combine statistical physics with theory of open quantum systems >20yrs ago, but my research is on statistical methods for biology. The talk will be on effective practices in computational biology and supporting tools & communities.
@mbeisen @BorisBarbour Yes - writing papers is needed to record scientific activity, but from readers’ point of view, we need better search tools, reviews, comparative analyses, aggregator databases etc. to help them find what they need to read or want to reuse.
@RArgelaguet @iscb_scs @embl @iscb @iscbsc @GonzaParra_ @CuypersWim @cuypersb @CleidyOsorio @VictorGrentz @caicebalt @KhalGabbo @Mina_2912 @krmsalazar @pradeeperanti https://t.co/akiHu5Tn5W
Subgroup-specific gene expression profiles and mixed epistasis in chronic lymphocytic leukemia https://t.co/3OY1Czsb6m (bioRχiv)
Thank you @Almut30618742 @ViktorLu_1118 @ThorstenZenz and Sascha Dietrich for this great collaboration.
@ZmbhH @kaessmannlab Congratulations Henrik! Looking forward to more of the great science you’re doing.
Big construction on the @EMBL campus. While the Imaging Centre is getting finalized and GSK Cellzome have moved into their shiny new building, the old container-building is being dismantled. https://t.co/gEvy06cce8
@BioMickWatson Just use the bibtex R-package on the bibtex file of your publications.
Explore and search the code of all of @Bioconductor https://t.co/Lq3Wv5odLV
@grimbough @Bioconductor Brilliant work, Mike!
Women in Data Science 2021 – Perspectives in Industry and Academia - 18 June, 15:00-17:30 CEST
https://t.co/dqnJP07326
@Meristemania …and similar trend across many European countries, https://t.co/eCbYqvgZGs EU about 6 weeks behind US and 8 weeks behind UK, but slope now is very good. Mostly due to BioNTech/Pfizer. https://t.co/Eh5SBfYml4
And this is of course exactly how science should proceed 🙂
@dermitzakis @GSK Congratulations! Great news.
@DrAnneCarpenter @HFarooq22 @michaelhoffman @laurastephen It also means that even if you manage to stay sane, one is always reactive and never proactive. Which contradicts why we have these jobs.
@DrAnneCarpenter @HFarooq22 @michaelhoffman @laurastephen Somewhat related, advice from @birgit_kerber, successful IP lawyer colleague: “The best way to receive fewer emails is to write fewer emails.”
@StephenEglen Of course:
> dadjoke::dadjoke() It’s inappropriate to make a dad joke if you are not a dad. It’s a faux pa.
@ewanbirney Father’s wants to build an irobot but just for legos, with fischertechnik.
@aaronquinlan FWIW, there’s the Ranges infrastructure in @Bioconductor that attempts that, https://t.co/ZW5Y1y8wGQ
The good and bad things about academia in one single meme. https://t.co/OyS8T0IPlr
Workshop: Mapping the Landscape of Genetic Dependencies in Cancer 2021-05-28, 14:00-19:00 CEST via zoom hosted by @Michael_Boutros et al. Exciting speaker list incl. Jesse Boehm, Matt Garnett, Francisca Vazquez https://t.co/IAr3bgfTKX
Science writing tip: avoid adjectives that allude to quantity but are actually not quantitative (very, wide, large, …)
The CSAMA one-week intensive course “Statistical Data Analysis for Genome-Scale Biology” is back! Next one 19-24 June 2022. It had taken place every June or July 2004-19, and needed to be cancelled in 2020+21 due to the pandemic. https://t.co/dNGkIZ7YH9 #csama2022
Programme and registration will be available in early 2022.
What if project managers of consortia did not see it as their job to send as many emails as possible to consortium members, but as few as possible.
@notSoJunkDNA I agree. Most are fantastic, and really helpful. My comment was tongue in cheek and, I hope, not offensive to anyone in particular. Trying to make the point that lots of communication + discussion != productivity.
@siminaboca Congratulations, Simina! A great opportunity.
@harshameghadri @zielinski_cz Sorry to hear - 65% salary is not OK. Graduate student (PhD candidate) working conditions is an international problem though.
@harshameghadri @Stefan_T_Huber @zielinski_cz It is not OK. Indeed I had a similar deal (50%) for my PhD in physics in the 1990s. It made me take a side-job as a programmer in the hospital to afford a better apartment and travels; and leave academia. (The fact that I later reentered as a bioinformatician is another story.)
Science is organised by scientists for scientists, so it is baffling how bad some of the designs are. Such as that there is a linear (1-dim) order of authors in a paper that needs to be parsed for significance and type of contribution.
(Take movie credits for comparison.)
On this splendid summer morning, the two-year old has the best commute to @EMBL kinderhaus. Cows mooing and birds chirping in the background. https://t.co/Gy9DF57udo
Do you all have a folder called “purgatory” for email that you only want to deal with …. much later …., or are you normal.
@aemonten Groundbreaking.
@IanSudbery I think you are onto something (exaggerated personality cults, reward by prestige rather than $$, etc.). Although of course there is no someone, and the goals are bound to vary across a heterogeneous group.
@jjloverp You can suggest to them that you are able to do it in (insert time period that works for you), and decline otherwise.
Sad and stupid.
I had my first contact with UK on school trip to London at 18. Later went to more parts of UK for a longer holiday, came to study 1yr in Edinburgh, worked for 5yrs in Cambridge as a researcher, made many friends there. Such ties take time to build, & start small. https://t.co/IZmIT1cSef
I’m by far not the most important person at my institute nor the most communicative one - nevertheless our IT team tells I’m the one with the most email. Shattered.
@lawrennd Apparently all desktop IMAP clients known to humankind choke on it
@cwcyau The pendulum will swing back. The younger generation are alright.
Great talk yesterday by Dmitry Kobak @hippopedoid with many interesting and perhaps non-obvious insights into t-SNE, UMAP & Co. at the ELLIS Life Data Science Seminar, now on youtube https://t.co/VoiX1F9Hgv https://t.co/nAmT0fzMaz @ellislifehd
MatrixQCvis: shiny-based interactive data quality exploration for omics data - by Thomas Naake @tnaake7 https://t.co/2PCCB4Xh9P https://t.co/kwyPDj6PfP
Imagine there were no internet and each time one computer wanted to talk to another computer you would have to worry what lines there are, who owns them, and how to rent one….
@biocs Amazing but in accordance with the group_by semantics - the factor levels are assigned differently in the two cases as in one case the grouping needs to be honoured, in other it doesn’t. I agree this is quite subtle and cryptic. For robust code, always set levels explictly….
@biocs E g. Try with levels=as.character(1:5) or indeed with levels=as.character(-100:100)
First mountainbike ride in the Alps since the pandemic While the kid is having fun in the playground of the fabulous kinderhotel. https://t.co/VNZHOVzY0t
@biocs But then maybe better sort your data by numeric value (e.g. rank) rather than by lexicographic order?
@thomas_sandmann @const_ae @satijalab @NimwegenLab Thank you, Thomas, for the nice feedback!
Preprint by @const_ae on major types of variance stabilization and transformation approaches for single cell count data: delta method based, Pearson residuals, Bayesian latent state inference, comparing their interrelationships and relative strengths and drawbacks. https://t.co/h4oiBmlJvJ
For the record, URL/DOI is https://t.co/toiipUjhXQ
(I shouldn’t be tweeting and multitasking.)
@NimwegenLab @const_ae Hi Erik and co-authors, Sanity is very good, and we hope that this is clear in our paper. It is part of academic discussion to always try to look at both sides of a coin. Indeed we only talk about ‘potential downsides’, which is meant to be pretty light, and maybe (1/3)
@NimwegenLab @const_ae we can find a better word in subsequent versions. Your paper (title) makes a lot of of the Bayesian approach (which is great), and one of the beauties of this is having posteriors rather than just point estimates. In the case of Sanity, … (2/3)
@NimwegenLab @const_ae … the multivariate posterior has a rich correlation structure, so it is (inevitably) conceptually and aesthetically a bit unsatisfying to - as you suggest - just drop that and replace it by a single number, the expectation value of the posterior. (3/3)
The Protein Landscape of Chronic Lymphocytic Leukemia - with F. Meier-Abt, Junyan Lu (@ViktorLu_1118), R. Aebersold, @ThorstenZenz https://t.co/S2453Vv2Yu https://t.co/RhOXQqoqrW
Discovery of a new biological axis of heterogeneity in CLL using multiomics factor analysis, by Junyan Lu (@ViktorLu_1118) and Ester Cannizzaro https://t.co/fvVOnY9Okm https://t.co/xZyGIARh49 https://t.co/P2850ReniE
The analyses presented in the paper are third-party reproducible: https://t.co/bJGfOlreLi
https://t.co/pHo8K1edrk
@ewanbirney In the beginning, there were positive integers. Then people wanted to solve polynomial equations, like a0 + a1x + a2x^2 + a3 * x^3 + … = 0. So they successively realized they needed to introduce negative integers, rational numbers, irrational, imaginary. And then it stopped.
@ewanbirney Yup, there are multiple approaches. The algebraic, where numbers are just things with which you can do algebra. (v. abstract) The geometric, where you picture them as points in some space. The physical, where you want that space to match something real.
Great news you are coming to Lausanne / Switzerland / Europe! Wish you a great start. https://t.co/fuXa3nBYtH
@lawrennd Whether your premise is true is hard to judge without data; perceptions are subjective. And efforts like DORA are getting more traction that point out that reputation of a journal (whatever that is) is not the same as the quality of contained articles.
@lawrennd Maybe these publishers are indeed asset stripping while they still can…
@tobiasgalla Photos are not required, expected or helpful for applications at @EMBL.
A nice example of interdisciplinary collaboration in precision medicine with researchers from EMBL, Univ. Hospitals Zürich and Heidelberg, DKFZ, Cambridge, Barcelona, Erlangen, OSU highlights the leading role machine learning can take in ’omics. https://t.co/ppegIiFMfb https://t.co/LzXo3oC4yP
@jamie_daw Is it not an example of a “the rich get richer” system that key players (who are actually not the government) have an interest in maintaining?
OLISSIPO Inaugural Workshop on Computational Cell Biology 20 July 2021 (14-18h CEST / 8-12h EDT) Four exciting keynote speakers: Susan Holmes (Stanford) Dana Pe’er (MSKCC) Luay K. Nakhleh (Rice) Rolf Backofen (Freiburg) Register until 18 July! https://t.co/TekN2LVtHU https://t.co/mJ0b3gvSsC
@larsplus Well the better answer is that research software should be better engineered - which needs processes for evolving a piece of code from research output to reusable product, dedicated engineers and funding. @Bioconductor is one effort in that direction.
@KLdivergence I bike commute, which luckily is jusy a short steep forest path (220m altitude gain) from Heidelberg downtown to EMBL. And sometimes a few detours - like now, Friday afternoon hangout on forest playground. https://t.co/PKKYLHedFs
@KLdivergence Doesn’t come without challenges. Like changing nappies in the field.
@JoanaFFPViana I guess you’ve seen this…: https://t.co/BStLquXxn9
@iscb_scs @y_bromberg @Nicky_Mulder @CleidyOsorio @GonzaParra_ @pradeeperanti @CuypersWim @cuypersb @GabosLab @Mina_2912 @propicee @iscb @iscbsc The PDF file of my talk slides https://t.co/82w2LVGVLP
@iscb_scs @cehijar @iscb Thank you! This is so cool. I am very grateful for the opportunity to talk at the conference.
We experimentally tested how different microenvironmental signals modulate the drug responses of CLL & how that depends on genetics. By Holly Giles (@hollygiles96) & Peter M Bruch via the Molecular Medicine Partnership Unit of EMBL and Uniklinik HD: https://t.co/VwLZfcwBCy https://t.co/la54fkMrvi
The computational analyses in the paper are third-party reproducible: https://t.co/d4rjkU4qCD
Exciting opportunity - biotechnology development with application to tissue biology and genetics https://t.co/4zHsxZ0UP7
@larsjuhljensen You can play with the data here: https://t.co/eCbYqvgZGs
None of these countries have saturated; and e.g. Spain has similar vaccinated numbers as UK.
I feel seen. https://t.co/IyC6Zr6eFO
@florianjug I started to look at it as a glass half-full vs half-empty thing: worry not about all the things you didn’t manage, be happy about those you did.
Also, many tasks that come in by mail are reactive, and it’s good to balance the time for that with proactive work.
1/ In this tweetorial, I present how multiple testing can be made more powerful by using freely available informative side information (“covariate-powered multiple testing”), as described in a new paper @NikosIgnatiadis (https://t.co/TErBj6elgp).
2/ What a scientist typically cares about when they need to decide whether to publish a certain claim is “what is the probability that, if I make such a claim, it’ll later turn out to be wrong, and I’ll be embarrassed.” In statistics, this type of notion is implemented by …
3/ the false discovery rate (FDR). In the language of hypothesis testing, a ‘rejection’ is synonymous with ‘discovery’, but it’s important to realize that an FDR has no easy relation to a p-value. It’s possible for the FDR to be large even if the p-value is small—for instance…
4/ if you found your discovery by chasing small p-values among millions of hypothesis tests. On the other hand, the FDR can be small even if p is large—e.g., if the statement is likely true a priori anyway, and you just picked a bad experiment or had bad luck executing it.
5/ It was a key insight by Benjamini and Hochberg (BH), published in 1995 (https://t.co/ZZglBQRRml), that it is possible to link p-values and FDR by doing a “multiple testing correction” if one is willing to work with the worst-case upper-limit that …
6/ all discoveries are wrong (all hypotheses true), and to ignore any thought of a priori probabilities. Storey et al. (2002; https://t.co/v1C3Q4G2gW) modified BH by bringing a priori probabilities into the game, but their method assumed that they are all the same. …
7/ In real data, not all hypotheses are created equal, some are a priori more likely, and some may have better (more powerful) or worse (less powerful) data than others. In bioinformatics circles, this kind of folklore was reflected in ‘prefiltering’ (https://t.co/rt7Je61zPg).
8/ But dichotomizing a continuum is seldom the ultimate solution, so several groups began toying with using side information on hypotheses in a more gradual manner to guide FDR control. Our first stab at this was Independent Hypothesis Weighting (IHW; https://t.co/NKoewnnIyy).
9/ Its premise was that in a multiple testing setup, domain scientists usually have additional data that are informative about prior probability and/or power of each test. The information is often indirect, but nevertheless it can be exploited. The key result was that …
10/ it’s not the best strategy to just rank all hypotheses by p-value and pick the top of the list. Instead, it’s better to multiply each p-value with a weight factor that amalgamates test-specific prior probability and power. Thus, sometimes a mediocre p-value … https://t.co/AkNVxWlmHx
11/ for a hypothesis with a high prior probability of being false could trump a small p-value of one with a lower prior.
There is, however, the important concern of overfitting these weight factors. If this happens, then p-values are artificially inflated, …
12/ we lose type-I error control, and the whole idea goes up in smoke. To avoid this, the 2016 paper proposed three measures: convex relaxation, cross-weighting (a form of data splitting), and regularization. The main contribution of the current paper (https://t.co/TErBj6elgp)..
13/ is to disentangle the incidental from the essential. It turns out that cross-weighting is the key idea: train the weights on a different subset of hypotheses than where you apply them. The method can be applied—in a kind of ‘modular’ manner—to different …
14/(END) multiple testing methods. In many applications, it increases the power of multiple testing methods essentially for free. https://t.co/RijkaQyx6n
This work has also been presented, in various stages of evolution, in arXiv: https://t.co/jnGjxv7eHk
(Btw, personally I prefer the text & formulae layout of the latest version on arXiv to that in the journal.)
Group Leader positions in Theoretical Biology at @EMBL: https://t.co/sBqWqUzFwM https://t.co/lamDnA6Ash
@wblau Also in the equally sunny state of Baden-Württemberg: https://t.co/ePfwIfqnq8
@KevinRUE67 Not quite what you are asking for, but here some simulations and visualisations on why you’re question is a good one and why the answer is hard https://t.co/2hmCryROly
Perhaps the approach @daniela_witten presented at #bioc2021 could be useful.
@ewanbirney Sounds like bioinformatics, except that there is never any units and it’s called batch effect correction.
@Frutag33 At @EMBL, the employment contract is between graduate student and institution, for at least 3.5 yrs https://t.co/osxUZE61X1 If for any reason a student can no longer work with a PI, the graduate office and the thesis advisory committee work to find an alternative solution.
Research software engineering is so centrally important - and it needs to become a more tangible, more attractive career track https://t.co/nxUWJRCTQD
What is your mirror event (birthday - age)?
The geodesics from my birthday to me tweeting are now longer than those from the publication of Einstein’s general theory of relativity to my b’day.
An email account where incoming messages are ranked and given visibility depending on donations to a charitable cause by those that write.
One of the uplifting scenes this week, besides the award of Germany’s Order of Merit to former EMBL director Iain Mattaj, was to see the republic’s representative, minister @TheresiaBauer, who presented the award, come up to EMBL via Steigerweg on her e-bike. https://t.co/FNDVfLJU19
@OliverStegle @BenLehner The years in which scientists are most productive (at least, directly) usually precede those when they are most engaged in power politics. So not sure the link between competition and productivity is inseparable
@OliverStegle @BenLehner There is a pre-existing objective function (discover truths about reality, develop new tools etc) that’s independent of winning competitions.
Ana Teresa Freitas and Mike Inouye @minouye271 on clinical utility of polygenic risk scores. This Thursday 14:00 CEST (Paris/Berlin/Rome) online. https://t.co/WV1o3Mt7mi
Congratulations, and best wishes for good luck and continued success to Katharina! She has been a joint postdoc between Michael Boutros and myself and been a fantastic colleague and a pleasure to work with. https://t.co/gRZyKaArGy
In memory of Clive Sinclair. ZX81 with 1kB RAM was my first computer. I learned Z80 assembly with it and many programming tricks by studying its 8kB ROM OS. As a 14-year old I had been reading the book by @RolfDieterKlein and … (1/2) https://t.co/8QB9oPGsxl
… wanted to solder together something along those lines from scratch. When I sent him a postcard with a question, he gracefully replied and advised me to save up and buy the ZX81 instead 😌. https://t.co/tL2CVMZPso
@larsjuhljensen Just an accident of when one was born, I guess. Someone here will have manually programmed punch cards.
NCT Data Science Seminar on Data Journalism with Marie-Louise Timcke, Wed 2021-09-22 11:00 CEST. https://t.co/Q4SzLGYtS2 (incl. registration link) https://t.co/nb8ckNytwR
@zhisong_he I understand the sentiment, but I think it’s only partially true. Many good scientists understand the importance of those who do or enable the actual work very well. As for those who get wound up on titles and prestige, they probably miss something else in their life.
@fepinheiromycin @humantechnopole Congratulations, Fernanda, great news for @humantechnopole and for you!
The ICE train from Mannheim to Paris is such an example of differential investment in infrastructure. Up to German-French border, dawdling along at 110km/h jumping & shaking on bumpy track, then after Forbach, accelerate to 320km/h for a smooth ride across beautiful countryside.
… my first business trip since December 2019. So excited to join @EMBL faculty meeting in Paris.
@BrenRouseHD The train was great: comfortable seat, fast WiFi, got plenty of work done, picked up a coffee & croissant from the bistro, nice views from the window, and then a pleasant walk from train station to venue through Parisian streets with sidewalk cafes and fashionable people.
@AedinCulhane @MedicineAtUL @RTENewsNow @RuthClifford5 Congratulations Aedin - an exciting and important new challenge!
Attending a hybrid conference, and the variability of sound quality of the remote participants broadcasting into the meeting hall is staggering. My advice: buy an affordable semi-professional condenser microphone, rather than relying on the built-in tinbox thingy in your laptop.
@IAmSamFin Yes — see https://t.co/qMJYoFTwma or indeed https://t.co/iidRBAf0Px I use the Rode NT-USB.
Goodhart’s law: When a measure becomes a target, it ceases to be a good measure. https://t.co/HQVqJNbu3E
Smita Krishnaswamy Geometric & Topological Approaches to Representation Learning in Biomedical Data Today 15:00 CEST, 9:00 EDT Open to all, join via https://t.co/L9T4gHA0uJ https://t.co/Du1HgolK2t
Gave a talk yesterday at the (virtual) Uni Luxembourg Life Science PhD Days and received this lovely present for the poster session. Thank you to the organisers - Apurva, Céline, Eleftheria, Kristopher & Mina. Hope to visit in person some time soon. https://t.co/VRwz3wv4pm https://t.co/gezlFq2PpH
A short write-up arguing that the primal concept in teaching hypothesis testing should be the false discovery rate. In single testing, the FDR is often not knowable, while one can compute the p-value. Which is better than nothing, but the right answer to the wrong question. (1/2) https://t.co/oBUqpJEhpK
FDR and p-value are vaguely related, but not even monotonically. The great thing about multiple testing is that the answer to the right question, the FDR, does become more accessible. https://t.co/qE6KwDVNaK
If Facebook is the Big Tobacco of the 21st century, then maybe our grandchildren will look back ať our current social media use like we look at the smoking habits of mid-20th century people?
@strnr For mountains, the Dolomites including https://t.co/1JuCqajONO and Cortina d’Ampezzo. Also great hospitality, refuges with good food, …
85% of statistical consulting is helping people figure out what the right question is, 14% is data wrangling, and 1% mathematical methodology.
(Saying this with no disrespect for methodology - I love it…)
… and to qualify, since this sounds a little harsher than it should: One needs to know the mathematical methodology in order to even be able to propose what the answerable questions (and caveats) are.
Analysis by Dmitry Kobak @hippopedoid and S.Shpilkin on statistical patterns proving election fraud in Russia reported by the Economist. https://t.co/FpoXe8Sorl
The never-ending quest for the right denominators (or ‘normalization’): https://t.co/i6E3GXpolm
Great opportunity in Manchester https://t.co/qyGT9L66tk
@fabian_theis @PhilippWeiler7 I think the processes of big open source projects such as Bioconductor, incl. compartmentalization (packages/plug-ins), interoperability, life cycle management, support forum, continuous integration are crucial. Also, hire research software engineer(s) if you can.
Ascona Workshop 2022: 27 March - 1 April. Biological systems: from first principles to data-driven modelling and back. https://t.co/QuibKrQbOV Preregister now. https://t.co/ovG4ppoPLu
Group retreat 2021 in Niedersteinbach / Alsace with lots of in-person discussions, amazing food and hiking. https://t.co/QKaT9ZLA20
@michaelhoffman Bioinformatics analyses need positive and negative controls, just like molecular biology experiments.
@michaelhoffman It is more comforting to do an expansive methods benchmark than to try to use anyone method to discover a new biological phenomenon.
@BioMickWatson You could look at the plots provided by the MatrixQCvis package on Bioconductor
This. Without automated choice of all parameters, it’s not a good method. https://t.co/DhYFUvZbaU
@vallens @michaelhoffman Without Bioconductor it would be thousands
(albeit with a different definition of “do”).
The first and the last sentence of your paper’s abstract should not be the same.
The difference signals the advance that was made through this work.
@OurWorldInData Where high street drug stores sell healing crystals and pharmacies peddle homeopathy.
One used to be able to think of this as a harmless stupidity tax, but now it turns out to be a problem.
@mjskay Why stop there
library(“magrittr”) =(y, 9) %>% :(1, .) %>% for(x, ., {( {(*(x,y) %>% cat(x,“*“, y,”is”, ., “”)) %>% if(x>2, .) )) https://t.co/TlCo5yOVao
@deepfriar @mjskay Boss level R
@fabian_theis @kwbroman Nice analogy. Also, physicists doing mathematics can be different from mathematicians doing mathematics.
Very interesting effort at a peer-reviewed journal for reproducible scientific articles (literate programming, Rmarkdown, JuPyteR notebook)!
I’m interested in learning more, e.g. how to deal with updates/bugfixes/changes in dependencies, and with non-standard compute resources. https://t.co/veaKmUQ9Rp
Privilege and prestige in academia.
These numbers are from US, curious how it looks in other countries (expectation: similar). In Germany, there’s a drive to increase the role of prestige with the Excellence Initiative (https://t.co/FNqd8xjTik) as a response to …(1/2) https://t.co/25DpolGGxj
perceived weaknesses of the traditionally more egalitarian state-run system.
As someone from a family of farmers & factory workers, where no-one went to college, I feel lucky. Some of this is thanks to the sweep of social democracy and egalitarianism in 1970s Europe…
There is privilege of socio-economic class, and there is privilege of country/society you’re born into. Both should play little role if the goal is to get the best minds and talents into a profession. (Sadly, current historical tides go the other way.)
@profwehler Surveys are used by committees that have no inherent authority on a topic to gain legitimacy by selectively employing an element of democratic governance.
Hi! Ich heiße Wolfgang Huber, bin Bioinformatiker und Statistiker für biologische Daten. Ich und mein gesamtes Team sind geimpft, weil wir gesund bleiben wollen. Wissenschaft & Solidarität sind der Weg aus der Pandemie. Deshalb: Lasst Euch impfen! #allesindenArm
The French pro-vaccination campaign vs the German one https://t.co/dpD6bvR6pM
@ManuelKaulich @TraverHart @Michael_Boutros Manuel, can you provide the zoom link in clickable format?
@leonardocarella @chrishanretty Not sure if that’s what you’re after, but if you resample from an infinite pool, all is fine. The bias in your bootstrap estimate sample_means_1 comes from correlations induced by always sampling from the same 10000 observations. https://t.co/3oEiDMyUks
Starting now https://t.co/y0VMTpBA7y
Rule #1 of academic management: a PI’s time comes for free, so please put as many meetings, surveys, committees, reports, reviews, quick requests to double-check or comment on them as you like.
Being a scientist https://t.co/eOGZmCMSOG
@AedinCulhane @Bioconductor @OHDSI All of these!
How to think about method development in computational biology https://t.co/kH8be0ytqH
@BoulesteixLaure I see your point about conflation & agree it’s sometimes a problem. OTOH, building a new tool & then not being able to do something one couldn’t do before also seems unsatisfactory. (If authors themselves can’t be bothered, why shld anyone else?) As always, optimum is in-between.
@mikelove @BoulesteixLaure Agree, developing a new method is a lot more than showing a one-off lucky application to a new data set. Application examples are important, but robust software, documentation, maintenance etc are what makes it useful and sustainable.
@IanSudbery I’d start looking at learnr from Rstudio and the exams package on CRAN - perhaps this is programmable enough to do what you need?
@mbeisen FWIW, the same statement in 2 pages: https://t.co/OoKIPjKbag
@drisso1893 @mbeisen Attempts at humour in scientific publications should be discouraged.
(And yes for a while I did ask for the definition of the p value in interviews of applicants who claimed a statistics background, and you wouldn’t believe it.)
TFW you feel the urge to include a reference to ISO 80000-2 Part 2 Section 3 every time when editing a lab member’s manuscript draft.
(It states where to use italic type or not in mathematical expressions.)
On the way back from a most beautiful family week in Tenerife, to wintery Heidelberg and a busy (but mostly fun) schedule of work. https://t.co/CzgX4b1faO
@Karl_Lauterbach Herzlichen Glueckwunsch! Es ist gut, dass Sie mit Ihrer Fachkompetenz und Klarsicht dieses Amt uebernehmen.
Today 16:00 CET Helen Parkinson from EMBL-EBI https://t.co/LoLpNw1iN5
The cool and efficient way to work with data along genomic coordinates. https://t.co/A7v6KnFEvt
Good thread by Moritz on the situation and outlook in Germany. https://t.co/XLO1iJw1pv
@markowetzlab Sorry to hear. That is so intrusive.
Sad to hear about the passing away of Günther Sawitzki on 25 Dec 2021. He was a pioneer of statistical computing. https://t.co/N4WZEr6idV https://t.co/ZYmCF84GFE
Among many other things, Günther organized the SoftStat workshop series, and there in 1995, R made its first general appearance, with R.Ihaka and R.Gentleman coming over to Heidelberg from NZ - leading to CRAN, R Core etc.
Good advice for 2022. A happy and healthy new year to all of you! https://t.co/AOgrmI7miW
Being busy ≠ being creative or productive. Two pieces to help put email, slack and zoom in their place: Stand up to your bullying inbox https://t.co/zcEQWMqOxO The rise of performative work https://t.co/DWsCpSxP9P
@anshulkundaje @mikelove @stephaniehicks @SherlockpHolmes AfaIu it’s a regular setup for t-test (or Wilcox, KS if you must). As Susan says, size imbalance is not a problem (convince yourself doing simulations on synthetic data). Things to check are: - are the 2 distrib’s reasonably unimodal - independence(!), esp. for the smaller group
@anshulkundaje @mikelove @stephaniehicks @SherlockpHolmes … and in your case, from what I understand, independence is given if the loci are unlinked. (If they are, reduce to a s subset of unlinked ones.)
@anshulkundaje @mikelove @stephaniehicks @SherlockpHolmes @SherlockpHolmes ’s suggestion is about testing whether the allele frequencies differ between the two groups, mine, whether the predictions differ.
Organizing a scientific meeting. Last year: let’s move everything online. This year: let’s wait for better weather.
How to estimate protein dependencies of patient derived tumor samples, based on - tumor x drugs response matrix - drugs x proteins binding matrix https://t.co/ZvvCmdSnW8 https://t.co/wm8WEmNIO9
We have two exciting PhD level Research Software Engineer / Data Science positions in medical AI at EMBL via the Heidelberg-Mannheim AI Health Innovation Cluster. Jobs: https://t.co/goDK7CxEaR Context: https://t.co/7OdRVaGJuq (1/4) https://t.co/U6jD15taDF
- Research Software Engineer in genomic and multi-omic data science, with Oliver Stegle (@StatGenomics), to work on new interfaces between R/Bioconductor and human omics database infrastructures such as the German Human Genome-Phenome Archive (https://t.co/vx3wCxTkT8) (2/4)
- Data manager | Research Software Engineer in precision oncology using single cell multi-omics and multiparametric imaging, to assemble and curate large datasets from clinical sample cohorts, and as part of the team, mine them. This is … (3/4)
… with @DietrichLab from hematology-oncology https://t.co/n5VFrsEFi3 Interested? Please get in touch. Deadline: 2022-01-31 ! (4/4)
Don’t want to be too melodramatic, but this trend of billionaires-funded science seems in line with the broader rise of authoritarianism around the world. Of course democracy is the worst form of governance, except for all the others. https://t.co/bwiphiPk0n
@B_Esteve_Altava Difficult questions. Two exhibits: - There are many societies with super-rich individuals and meagre science systems. - The superefficient system in the US since the 1940s has been government-built. (Also, afaIu, those in UK, F, DE et al. in 19th century.)
Just read this in a postdoc’s career development plan and it’s perfect:
“…From the soft-skill perspective, I would like to improve my abilities how to maintain current and establish new collaborations most efficiently without unwanted and ineffective commitments.”
@dagarfield Yep. There are other examples of privatizing what gov’ts used to do, around the world -health care -education (prim-,second-,tertiary) -water, electricity, waste -law enforcement -law making -defense Diff’t people will have diff’t views on how well it worked out in diff’t places
@mikelove @nomad421 @daniel_c0deb0t Very much agree! See e.g. the approach taken in Modern Statistics for Modern Biology https://t.co/obiejJIy4R with @SherlockpHolmes Visualization, EDA, discovery of patterns in the data and a more general understanding of uncertainty take priority over tests.
@B_Esteve_Altava Not sure where you are, but here are some playgrounds in HD with “always” many kids: Synagogue, Danteplatz, Fire station, Farm & Gadamerplatz along Bahnstadt promenade, Neckarwiese
I did 1990-91 at University of Edinburgh as a physics undergraduate from Freiburg / DE. Learned general relativity from Peter Higgs, meteorology and weather forecasting, Fortran programming, and above all, fantastic people, highland hikes, and parties. https://t.co/WM8WehHThq
We’ve an opening for a postdoc in computational methods development (3yrs+). Looking for someone with background in higher maths (lin alg, probability, calculus & geometry), good computing skills, and interest in new biotechnologies & biological discovery. https://t.co/JRfDmpJuFr https://t.co/jUL3Ovd98m
The project descriptions for these two staff scientist (research software engineer, data manager) positions are now online, as well as several exciting others: https://t.co/dRRH7Xs8at
Apply by 31 Jan via https://t.co/iYpto12H4v https://t.co/i1DOgtRf9x
Journalism is often based on anecdotes, samples, storytelling. Glad to see this recent trend towards ‘big data’ & statistics-driven analysis in the media. A trivial, but fun example is this cluster analysis / heatmap of local music tastes based on Spotify. https://t.co/aAegK2atSr
Here’s another example of data journalism, in the Financial Times, on a much more serious topic. https://t.co/yW3XipsPIh “Full reproducible R code for this story is available on GitHub”
Email is the new Fax.
Ouch… “Paradoxically peer review, which is at the heart of science, is faith-based, not evidence-based.” “…slow, expensive, inefficient, poor at detecting errors or fraud, prone to bias and something of a lottery”
by a former editor of BMJ https://t.co/yIQVvo3YaT
Comp-bio GL job in a great new institute in one of the best cities in the world. https://t.co/Nm8W9eSmy7
If you think cutting-edge AI and ML in biology and health is hard. User authentication for resources needed across multiple institutes is harder (IMAP rules…)
@NimwegenLab If scientists don’t have the ambition to solve a hard problem, who would? IMHO, peer review per se is a good thing, what’s problematic are the gatekeeper & certification roles it’s been given. Post-publication peer review seems promising (with important questions remaining).
@ZavolanLab @NimwegenLab I like your analogy with news. Indeed the decentralization & laissez-faire of social media created a huge fake news problem. OTOH, traditional top-down media also had(have) blind spots, could be inefficient & are easily abused in some societies. Would we really want to go back?
I’m ashamed and angry about the naïveté &complacency of successive German governments in their attempts to appease Putin &his system. With the support of much of German society. With catastrophic consequences, and the country now unfit to even head-on confront the dictatorship…
There are buddings of a fundamental reorientation across the political spectrum, which I welcome. The Russian people are so much better than Putin’s murderous regime, and hopefully there will soon be a time for atonement. Victory and peace to Ukraine.
@felbalazard Nuclear is only one of several options to replace gas & fossils, and as far as I am informed, neither the cheapest nor the safest. Renewables, too, were neglected in the decade since Fukushima.
For the record, a lot has changed in the last 36h, e.g. https://t.co/llo6GehQwm
CSAMA, the summer school on Statistical Data Analysis for Genome Scale Biology with R/Bioconductor is back! Brixen/Bressanone 19-25 June 2022
(1/2) https://t.co/cDRyRBLOVS
Faculty: Laurent Gatto • Robert Gentleman • Wolfgang Huber • Katharina Imkeller • Martin Morgan • Johannes Rainer • Davide Risso • Lori Shepherd • Charlotte Soneson • Levi Waldron
Registration to open soon. Enquiries: bell(at)embl(dot)de (2/2)
New preprint on PhD and postdoc training outcomes at @EMBL, analysing the changing career paths for life scientists over the last 25 years. https://t.co/M4rJyd5IH9
with @JunyanLu1118, @BrittaVelten, R.Coulthard-Graf https://t.co/DJSwpk87vd
So when you upload a DOCX file to @biorxivpreprint, their conversion to PDF pixelizes the images with coarse resolution and compression artefacts (jpeg?) - whereas if you submit the PDF, you keep control over that (i.e. you can supply figures as vector graphics or highres bitmap)
Thread: a study of career outcomes of 2284 EMBL PhD students and postdocs over two and a half decades. https://t.co/QI9ortNMK4
@giorgiogilestro @embl @JunyanLu1118 @BrittaVelten Yes.
Prediction: Western democracies (incl. DE,UK,US,F) are just at the begin of understanding of how many political events of the last years (and politicians) were influenced by Putin’s corruption. And how to defend against such meddling in the future w/o sacrificing core values.
@SilasKieser @BioMickWatson https://t.co/x38yjVtMaF next to Feuerwehrspielplatz. Comparable setups at Bauernhofspielplatz, Zollhofgarten und Neckarwiese.
Congratulations, @Prof_Lundberg Well-deserved! https://t.co/jAFXNq0FW1
Positions at Luxembourg Institute of Health for Ukrainian scientists https://t.co/1hSUgHBKtY
@SvobodaLab I’d be happy to offer one- or two-week compact courses introductory statistics, data science, bioinformatics.
There’s an exciting inter-institutional EMBL/DKFZ postdoc position with @AurelieErnst and myself on machine learning on histopathology slides to detect clinically important genomic aberrations. See https://t.co/NGNEyRuk3z for more info.
The International Society for Computational Biology (ISCB) bestows one of its highest scientific honors, ISCB Fellow, to Mikhail Gelfand, Ph.D., for his pioneering contributions to bioinformatics research. https://t.co/tJAceK2fIf https://t.co/yLZ8xC5dHp
A thread on differential expression testing with large sample sizes. The null hypothesis of popular methods such as limma, edgeR, DESeq2 etc. is that there is absolutely no difference between the two groups (or, in more general designs, no significant effect in a linear model) /1
It’s well-known (and obvious) that this null hypothesis is, strictly speaking, almost always wrong. If you just look close enough, practically every gene is ‘differentially expressed’. For example, this plot from a 2001 paper by Boer et al. (DOI 10.1101/gr.184501). /2 https://t.co/cDPi82dR4u
When you take two sufficiently large sets of humans, there are always (possibly tiny) differences between them. Maybe the sex balance is not exactly 50:50, the ages are slightly different, or there is genetic structure. This translates into (tiny) differences between the … /3
Thus, only genes with more-than-threshold effect size are selected. As sample size increases, the set of genes selected as “differentially expressed” stabilizes, rather than just keep growing. Which is what you want. /5
averages of the expression levels of basically any gene. This is discussed eg in the DESeq2 paper. The proposed solution is to use a banded hypothesis test,where the null hypothesis is not exact equality, but that the difference is below a threshold. DOI 10.1186/s13059-014-0550-8 https://t.co/ZjDkfh29hy
Yes, this is a subtweet thread :)
Others have noted the problem of using a point like null hypothesis in the context of “too much power”, too. It has been proposed to fix it by using a test with less power. Maybe this works empirically in some cases, but conceptually, it seems odd, and poorly generalizable. /6
@AndrewHolding LogFC thresholds are important, but rather than applying one post hoc after test against a banded null, you can build it in upfront.
@holmrenser The issue is not whether the test is parametric or not, but the nature of the null hypothesis (point-like vs banded).
Permutation-based approaches have been proposed, but they’re complicated if not intractable if you have more than two groups or more than one covariate.
@AndrewHolding See the DESeq2 vignette Section “Tests of log2 fold change above or below a threshold” https://t.co/AGu6PJUexd and the section about this in the DESeq2 paper.
Congratulations, Judith! @zauggj Great to see this exciting line of research expanding. https://t.co/cmx7yiCxaJ
Pleased to learn that @EMBL-EBI is getting a 3rd building on the Wellcome Genome Campus, and that it’ll be named after Janet Thornton https://t.co/ikZ07omRCx
Exciting opportunity for mathematical modellers, physicists et al. on important topic with great and energetic new PI! https://t.co/N3ZhDdvqOx
Germans in particular might consider amounts that are a multiple of what they have given to the Russian government via their direct and indirect fossil fuel consumption.
A reminder that you can donate to the Ukrainian army: https://t.co/uqhJOeAtPo or https://t.co/pbO1VDJAIR
Petition to the German government to stop importing Russian oil and gas imports immediately (rather than within months, as is their current plan). It has already been signed by many public figures. https://t.co/1pEFxiyK2n
@JunyanLu1118 https://t.co/Z9Nzpb7cf0
CSAMA summer school 19-24 June 2022 Statistical Data Analysis for Genome Scale Biology in Brixen/Bressanone, Italian Alps
Registration is now open: https://t.co/8HILRjN56D https://t.co/Ktg8J8TAMh
Classes and labs combine underlying theoretical concepts, R/Bioconductor software infrastructure and hands-on work on real datasets https://t.co/8HILRjN56D 2/2
The CSAMA summer school teaches statistical and bioinformatic approaches to biological data: RNA-Seq and other sequencing based “multi-omics” assays, incl. single-cell and spatial. We also cover basics of mass spectrometry based proteomics and metabolomics… 1/2 https://t.co/fX2SIu3MyD
@merenbey Viz the sender-message-channel-receiver model of communication: all parties need to cooperate for a good result.
@brent_p @mikelove @LuciaBarMar The aim is to draw generalizable conclusions out of a specific experiment/study. For this, one needs measures of confidence in observed effects & estimates of variability. In good RNA-Seq data, technical variability << biological variability and … (1/2)
@brent_p @mikelove @LuciaBarMar … there is little lost in ignoring the former. One can (and should) however look at the individual technical replicates to see if this assumption holds. Deviations may indicate quality problems incl. annotation swaps, catastrophic machine failure etc. (2/2)
@brent_p @mikelove @LuciaBarMar All the common error models (incl. edgeR/DESeq2, limma::duplicateCorrelation) have a certain scope, but cannot deal with all possible sources of variation—such as sample swaps, reagent/machine failure. That’s why quality control, diagnostic plots, quality metrics are important.
Concise assessment of the current state of German government and industry leaders. Shameful. Polls indicate that the population wants oil and gas sanctions now. https://t.co/8g3CuEWL2s
@hippopedoid https://t.co/pe4Ss36241 also https://t.co/X5iwTSwFx7
@olgatokariuk Happy Birthday, Olga!
Congratulations, Christina! https://t.co/DFyYuCZcOs
Great tweetorial by @mikelove on analysing large datasets (big sample size, high statistical power) with methods such as DESeq2, edgeR, limma,…
(Btw I believe it’s a subtweet on https://t.co/Uo2KIeH2zt that ignores such basics and trumpets a non-effective alternative approach) https://t.co/7d9RSc4eqO
Great news. Congratulations, Barbara! https://t.co/d20Lgdyd8j
Well done, Johns Hopkins. And congratulations, Alexis! https://t.co/o8XJhrKplb
@larsjuhljensen Which 3D graphics device do you have in mind?
OTOH, by use of colour, faceting and perhaps shape one can accommodate 5-6 data dimensions (https://t.co/TdrfU31s6a )
@larsjuhljensen Sorry - the correct URL is https://t.co/zGrMsWqhSY (it seems Twitter swallowed the prefix).
Agree that VR should offer great new potential for data viz! But this field seems wide open, or at least I am not aware of much in statistics / data science applications.
@larsjuhljensen Yeah, 3D scatterplots are “obvious”, it’d be interesting to see new kinds of visualizations that make more use of our brains’ spatial orientation and spatial memory functionalities.
@larsjuhljensen Starting points could be arranging/sorting your data like items in supermarket aisles, or like shops, bars, alleys etc. in a downtown area, or like features of a landscape
@lena_maierhein That’s so nice. You must be proud 🙂 Some links are here: https://t.co/cNIGY7GpMU
@lena_maierhein … I wouldn’t have imagined saying this a few weeks ago: helping to ameliorate existing suffering is important and noble. If one wants to help avoid even more suffering in the future, giving to their army is the most effective means right now.
@leonardocarella Why, I have recordings of Русский военный корабль, иди нахуй, or of Ще не вмерла Україна. Or maybe Brahms again in better times.
@hallettmichael @jeffvierstra Re (1), if something is potentially useful for >1 person and is in R, put in a package and put that on Bioconductor, CRAN, or somewhere that essentially replicates what they do.
@hallettmichael I try to establish a culture in the lab where submit-ready manuscripts are accompanied by an Rmarkdown (or equiv) doc that reproduces all “results” figures, tables, etc. Some people may feel this slows them down, I think it’s the opposite (+ obv. reproduc’ty & quality benefits).
@kauralasoo @hallettmichael @nextflow Conceptually, it’s simple: raw data are what came out of a machine or what someone published. Computations shd be reproducible from there on. Of course caching intermediate results is fine. I realize complications from unique/expensive hardware & not wanting to waste electricity.
I’m offering CV coaching for war refugee scientists esp. from biosciences, bioinformatics, statistics etc. DM me.
“CV” here being shorthand for job application material and approach
Job offer: Administrative Assistant with project management responsibilities for the new @EMBL transversal themes on Infection Biology and Theory, located in Heidelberg DE or Cambridge UK. https://t.co/aJryx7KK00
Is overcommitteed a word?
@michaelhoffman @baym Many of those with influence over such matters have benefited and expect to keep benefiting from the current system.
When your actions and misjudgments have brought your country into a terrible position of weakness and timorousness, German government and industry leaders, is it not time to step aside, and leave it to the next generation?
Apropos the idea that rank-based methods are a panacea (e.g.for RNAseq data) an interesting blog post by @tslumley pointing out how they can lead to non-transitive, or otherwise undesirable, conclusions https://t.co/CIZ4hUWtvH
@larsjuhljensen I disagree. Parametric t-test usually works surprisingly well even for data that are not normal distributed (try ‘breaking it’ by simulation). Where one needs to be careful is when measurement errors are correlated (aka batch effects). Then, it breaks (as do many other tests).
@KharchenkoLab @tslumley Two relevant papers afaIcs: [1] https://t.co/28dxmvayg5 [2] Characterising transitive two-sample tests, Lumley&Gillen (sci-hub…) -Real world examples may be rare, but [1] provides one -Tests that contrast a univariate summary statistic for each group are more straightforward
@larsjuhljensen No, I mean it for all sample sizes. Try it out by simulation (e.g. https://t.co/MXCqkCXEwi)
For small sample sizes, there is no alternative to parametric tests anyway.
@larsjuhljensen One can do all that, it remains that the real problem is often the correlated noise (measurement errors, batch effects); not the distributional assumptions of t- or ANOVA-tests. Simulations can be great to also model the correlations along with the testing.
@larsjuhljensen Sorry to be such a joy today… E.g. Benjamini-Hochberg is very robust against non-independence (i.e. correlations). Again, just try out with some simulations.
@Afelia Ich wünsche Ihnen alles Gute, Frau Weisband! Was Sie gerade tun, z.B. diese Woche bei M.Illner, ist so wichtig.
#CSAMA2022: Summer school on Statistical Data Analysis for Genome-Scale Biology in Brixen/Bressanone, Italy, 19-24 June 2022. Travel and registration stipends for scientists who were displaced by war. More under the “Registration” tab: https://t.co/8HILRjN56D
For those interested in Germany-Ukraine relations, an important talk by historian @TimothyDSnyder from 2017. Eerily predicting some of the things now going on in DE. https://t.co/ItzliHLcfZ
Men on bikes pointing at things https://t.co/xsizR4srSX
Does anyone still accept reviewing papers for journals “blindly” without first checking out the preprint?
@dominic_grun That’s an additional feature - no preprint, no review. But the most important feature is of course that I want to contribute to making interesting papers better, rather than spend lots of time on boring gatekeeping.
@markowetzlab Congratulations! 🍾
@fabian_theis Congratulations! Cool project. 🍾
@fabian_theis @dominic_grun I’m not asking someone to do something - that’s their decision. But I need to prioritize my finite reviewer capacity among a much larger pool of requests, and this is one criterion.
Women in Data Science - Perspectives in Industry and Academia 2022-05-18, 15:00-17:30 pm CEST (virtual)
Idea: sell old baby/toddler stuff on ebay classified ads and give proceeds to Ukrainian army https://t.co/pbO1VDJAIR
One of the hardest things in a scientific manuscript that grows over multiple months with multiple authors:
Use the same term for the same concept, and make sure each term only stands for one concept.
Also, the idea that lengthy explanations of why you did or did not do something are somehow more interesting than simply saying what you did and what the result was.
@BachmannRudi The problem with being a “large” country and that most media outlets are still organized by national boundaries.
@theosysbio @lgatt0 Good stuff.
Although I was referring to much more mundane things, like choice of reagents, specific analysis method, or of figure/plot layout…
Applications are open at the Graduate School (PhD programme) of Quantitative Biosciences Munich (QBM) https://t.co/uBzqPNFAAL https://t.co/9hmzbVqa1M
Congratulations, Hanno and Stefan! https://t.co/CMt8UsWxwa
For the German followers… After the recent cringe “open letter” stuff, there is now also a respectable one https://t.co/vYATjKY8qF
@WalhoutMarian I’m afraid the ‘conspicuous consumption’ of letter writers’ time is the real currency in this tradition (words are cheap).
I’m not defending or condoning the tradition, just noting what one is up to if one really wants to change it.
A powerful ‘investigative journalist’ thriller. Timely topic, too. https://t.co/UoI8TmaKm7
Amazing location and speaker list https://t.co/Liv7GKJm6a
So you can “fix” the problem of inflated discoveries due to confounders / batch effects by using a known-to-be-noisier version of the data. Nice shell game trick. https://t.co/6l8bIlM9pc
Abstract submission for European Bioconductor conference 2022 is open! 14-16 September in Heidelberg. https://t.co/gVMHdADcMi https://t.co/FrAGUs29mN
Looking forward to! Also Kyiv, Chernihiv, Donetsk. And Kherson, Yalta, Odesa. And more. https://t.co/1YiNwCAMn1
Dinner speculation on EMBL 2030: 7th host site in Ukraine focusing (e.g.) on plant biology
A virtual event on opportunities for Polish scientists for cooperation with @EMBL This is on 9 June, 9:00-12:30 (not 9 May as the tweet below says) https://t.co/8o3uSlZWI9
@KasperDHansen @timtriche @MTomasson and spatial and causal
A short thread on applying to PhD programmes. What I’m saying is based on my experience with the @EMBL PhD programme. Some of it may generalize to other programmes across Europe or elsewhere. https://t.co/UiHEs2AQPa
- Ranking and selection are a mix of absolute criteria (academic achievements etc.) and relative ones, i.e., which person could fit to which research group/area. For the latter, faculty will often electronically search the application database, so please make sure to include…
- All applications undergo systematic screening by a team of assessors from faculty. Volumes can be high (e.g. ~1000 per round at @EMBL), thus make sure your information is well-organized, easy to read, and the important facts are easy to find.
- It is fine to get in touch with faculty who are of potential interest to you. They are also just people and a short personal interaction (even if it’s only an email) can stand out over a mere database entry.
…the right keywords. This can also include names of PIs—but please without seeming arbitrary or closed-minded.
- All applications contain free-text elements (e.g., at EMBL we ask for your scientific interests and previous work). This is where you can stand out! Make sure the text is clear, logical, and well-written, and contains some non-obvious information relevant to you.
- If your work is computational, include examples of crafts(wo)manship: GitHub repo w/ own code, Rmarkdown/JuPyteR notebooks of projects etc. I like to see BSc&MSc theses or internship reports, as they tend to give a more unfiltered view on work style than multi-author papers.
- The number of excellent and promising applicants is always much higher than the number of places, and “the cut” is necessarily quite arbitrary and depends on minute circumstances, in other words, some luck. So if you don’t get selected, don’t take it personal….
- For better or worse, reference letters are important. They show that you could convince someone who knows you to spend some of their own time writing a letter for you. There are very legitimate reasons why not everyone can do this, but then please explain.
EMBL accepts applications in two rounds per year. The next one opens in mid-August, for interviews over the winter and start in 2023.
For anyone embarking on this endeavour: Good luck!
… apply at several places, and also don’t be afraid to reapply – to set of recruiting groups and their interests keep changing at every round.
If you want to argue that your high-throughput experiment worked because replicates are well-correlated, please do not provide a p-value for the null hypothesis of no correlation at all. Finding interest in that null hypothesis beams really low confidence in your data. (1/2)
More useful measures are, e.g., RMSD or correlation coefficient (possibly with confidence intervals). (2/2)
Where “noise” refers to variation between what you consider the same cell type/state, and “signal” to variation between cells that you want to consider different. Stabilizing signal-variance would in essence remove it and be rather counterproductive.
Another subtweet: Variance stabilization is often stated as the objective of transforming single-cell RNA-seq data.using, e.g., logarithm, sctransform. Important to keep in mind that you want to stabilize the variance of the noise, not that of the signal! (1/2)
In stochastic modelling, “noise” is not an inherent property of a system, but a catchall term for system variables the modeller has (or wants) to ignore. (The one known exception is quantum mechanics, where noise is inherent.) So, for instance, in one modelling scenario,… (3/5)
variation (residuals) from that. And so on. Thus, modelling assumptions such as Gamma-Poisson (Negative Binomial) and their parameters (such as dispersion) are highly application-dependent. (5/5)
you may choose to compare two cell populations A and B and consider any variation within A as noise. The next day, you may decide to look into A more closely and model it by discrete subpopulations; or by a continuous gradient. Then, the new noise is the remaining… (4/5)
@stephaniehicks @jhubiostat @JHUDataScience @JohnsHopkinsSPH Congratulations, & well done JHSPH.
@MiRo_SPD Sehr geehrter Herr Roth, ich schätze Ihre Arbeit sehr. Heute morgen im DLF haben Sie die Geschichte von einer angeblichen Absprache perpetuiert, die unter Verbündeten (&Ukraine) weitere Konsternation auslöst. Was geht vor? https://t.co/146D95zQYd
Registration for the European @Bioconductor conference 14-16 September is now open #EuroBioC2022 : https://t.co/GWmMGHbXGe https://t.co/RCIAsDQFVZ
Clustering methods are just heuristics for fitting a mixture model.
@kiran_r_patil You’re right, the “just” was wrong. One can think of some algorithms as a heuristic for fitting a formal mathematical model - but does not have to. Some algorithms are just that, algorithms.
My point in the OP was the usefulness of model-based thinking about clustering.
@carninci @BienkoMagda I think it’s often about more than wages: career perspective, work environment, time-limited contracts, the whole employer–employee relationship, what one might call “respect”. Academia needs to figure what its possible strengths are, what it can offer, and use that strategically
On the topic of academia vs industry: one of my most memorable anecdota is from a friend who became institute director at a famous well-funded German university. Head of admin tells them: “our job is to make sure you guys (i.e. the researchers) don’t do anything wrong” (1/2)
Explicitly no mention of getting things done, creativity or productivity. The researchers as a liability that just creates costs and risks, and needs to be contained.
Fortunately, attitudes are different in many other places. E.g. at fantastic @EMBL 🙂 (2/2)
A reminder of @grimbough’s cool https://t.co/leP8AXtwaP site to search and browse the source code of all @Bioconductor packages.
@minna_alander Germany, not enough.
The hard part in hybrid meetings is not video, camera or software, it is the room microphone(s).
TFW the effect of some perturbations is on a continuous range, noisily measured, & the perturbations are compared to each other via Venn diagrams of “significant” detections. Can be done,…but why? Such a needless conflation of stat. power, measurement noise & underlying reality
@tamas_schauer Yes, but gets messy once you have many more than two conditions or perturbations.
(There is, of course, this gem from the banana genome: https://t.co/dXkLqjCIaT )
So, what are better options? E.g., the Bland–Altman plot https://t.co/7NcTD7lJTJ If there are many more than two perturbations (or conditions), doing it pairwise becomes tedious, so you can compare each condition to the overall average or median.
Thread: A tool for annotating cell types using known markers https://t.co/a5LmBdugog
@science_wallet I’m all for discovery of new cell types or cell states - but isn’t it better to do this on top or on the background of what we know already—rather than pretending we know nothing and laboriously rediscovering the wheel?
@lgatt0 Here’s the link: https://t.co/8HILRjN56D
After the first morning of theory lectures, participants working away at the practicals https://t.co/YQ72ZQXawN
#CSAMA2022 - social event, a hike on Mt Plose to Rossalm at 2200m
All teaching materials are available at https://t.co/5XF2pP30b4 https://t.co/yOGobwz3Te
Great project: https://t.co/oeTe4sEeoq
The “Code Availability” section looks exemplary. https://t.co/guHaaLRhIN
It is not ideal, some things could be improved, but it’s worth remembering that about the EU. https://t.co/u3tdnx1HYq
@bctallis Among ambitious parents around the world, how many want to send their children for education (a) to the West, (b) to Russia, (c) to China? I think this is an interesting weathervane.
@bctallis 100% agree. Never meant to advocate resting on laurels - but point out how precious it is what we have and need to defend.
In this context, for anyone who hasn’t read it yet: https://t.co/JedXQ1yHNj
On corporate travel booking systems…. Sounds plausible https://t.co/Eti7Mmofjs
@fabian_theis @iclr_conf Not extraordinary, but bad for the field. Authors can make claims, occupy a concept/idea and prevent everyone else from working on it since it’d not be ‘novel’ anymore. Also, ‘scientific claims should be falsifiable’ (Popper) and from that angle, it’s not even science …
@fabian_theis @iclr_conf Well, I don’t think Popper’s falsifiability criterion is unmovable dogma. But it’s useful, and if a finding is really an unrepeatable “one-off” observation, I’d call that an anecdote or a case study and indeed wait until someone shows generalisability.
“On Tyranny” by @TimothyDSnyder is a book for these times. Insightful and to the point; both from US and European perspectives. Impressive illustrations by Nora Krug. Also makes a very good present for someone you love. https://t.co/wN2OkDhOtR https://t.co/JtTCGeMXRZ
@erlichya Great threat and overall analysis. Phrasing the difference between research in academia and industry via “lower/higher bar” is not fair - they have different objectives. Otherwise I agree: academia needs to reconsider what rewarding research output is - should be more … (1/2)
@erlichya than chasing for placing a PDF on certain websites; and how to reallocate academics’ time, who are increasingly just busy with reviewing and regulating each other and organizing stuff, to core functions.
@dagarfield @erlichya Yes, I’ve been lucky that some have found the software and datasets I contributed to useful. But my previous tweet should also have been more respectful of “PDFs”: there is of course great value in conceptual insight and basic research for which there is no obvious product..(1/2)
@dagarfield @erlichya …and private investment payoff, but which is of interest for society at large. Think understanding the origins of the universe, the tree of life on earth, or indeed research on delivering mRNA into cells in the 1990s. (2/2)
Theory@EMBL retreat, on implementing the new research direction (‘transversal theme’) of Theory at @EMBL, with @ErzbergerGroup presenting. https://t.co/WW4CX1tTmf
Programme: https://t.co/a0JXeugdmv
Interested in working as a research software engineer on R / Bioconductor / Open Science with me and my team at @EMBL? Apply here: https://t.co/JM6JIQFuqu
Topics: robustification& life-cycle management of research software, interoperability, APIs, virtualization/containerization, software usability, scientific developer support, platform integration (e.g. R—Julia—Python), interactive dashboards, living papers, reproducible research
@irileniaN I’ve had good experiences with - implementing features on request in open source packages for pharma that wanted to use them - doing data analysis of high throughput screens for biotech startups
@fabian_theis @EMBO Congratulations, Fabian! A great addition to the EMBO community.
I knew, but I think many Germans still do not realize the extent and gravity of this. Two well-to-do German colleagues report they’ve been sharing their house and kitchen with groups of Ukrainian refugees since February and how that has been going: very well. Touching stories.
Two-day faculty meeting with colleagues I hadn’t seen for a while. Finnish tells me how angry and disappointed many in Northern Europe are about Germany’s irresponsible and dangerous Russia & CEE politics of the last 15+ years and its meek and spineless behavior now…(1/2)
Looking forward to working with @DietrichLab, Karin Tarte, Camille Laurent, Claudio Tripodo& Peter Horvath on bispecific antibodies in lymphoma—using μenvironmental profiling to understand treatment response& resistance mechanisms https://t.co/Irt80CHPsg @transcanproject #BIALYMP
Professorship in molecular, cellular or systems biology in Basel. All candidates exploring fundamental questions in modern biology will be considered, ranging from the physics of life to the dynamics of multicellular systems. https://t.co/W1p9JAIp9Z
Entry-level course (Data Carpentry):
Introduction to genomic data analysis with R and Bioconductor
with Charlotte Soneson @CSoneson, Laurent Gatto @lgatt0 in Heidelberg 12-13 September. https://t.co/1qRv5vjBnK
Register via https://t.co/gVMHdADcMi https://t.co/WxeCWw1kUZ
A hoppy idea: https://t.co/ZW7NizRHLY
Good thread https://t.co/6hpULY6PRx
Congratulations, Mike! Great catch for @UNCBioStat https://t.co/VwjypuudvA
Job opening: Group Leader - Theory@EMBL @EMBL is recruiting a new group leader who develops theoretical approaches to understand principles of biological complexity, in interaction with modern experimental data types and computational approaches https://t.co/rsNFOpgash https://t.co/KFvD9cNlRM
Spatial statistics can help exploring spatial single cell data such as from multiplexed IHC or MERFISH. The MSMB book contains a gentle introduction into some basic concepts https://t.co/NmkS5qD36F https://t.co/BE3Yp4umRk
@nstroustrup1 ‘modern’ - not a major point, but…: similar as everywhere else (particle physics, cosmology, …), if there are data of better resolution, coverage, or more directly related to the underlying phenomenon, then wouldn’t a theorist prefer engaging with these?
Question: what is state of art and experiences with image viewers (zoom, move, select different color channels etc.) for multiplexed immunohistochemistry slides with, say, 60 Megapixels x 50 color channels, and many of these. R/Shiny app, EBImage, big-RAM server? Other options?
@K_Imkeller Thank you!
I was thinking of delivering QA/QC & result visualizations to collaborators via the web, through shiny & associated functions in EBImage: https://t.co/e5j4ETWwjD So for example library(“EBImage”) example(displayOutput) opens a shiny app with a basic image viewer.
@jokergoo_gu @HeidariElyas Can you give examples? I assume this primarily true for career development grants, while institutional recruitment to core- or project-funded positions wd usually be more flexible?
Such time limit on explicitly “junior” positions is a 2-sided sword—can also prevent exploitation
Leopoldina fellowships for postdoc level scientists from Ukraine affected by the war, to work in Germany https://t.co/jLc1UptGez https://t.co/V94VjHicpS
Twitter likes randomly cutting prefix info such as “www.” from URLs… Here is the correct one: https://t.co/H9yCiMQ48O
@GrigoriiNos Agreed. I’m not affiliated with Leopoldina or have insider knowledge, but from how I read the call, physically moving to DE is not required, it even seems written for cases such as you mention. Those interested should get in touch with the provided contact address directly.
. @EMBL has re-affirmed and summarized its commitment to good practices in research performance assessment: https://t.co/ssicVMyHZ4
(PS it’s been a great experience to co-lead the working group together with @jomcentyre )
Rstudio (the company) is becoming Posit. RStudio (the IDE) will keep that name. https://t.co/TMMWR79OmR
Full Professorship Neurooncological Bioinformatics in Heidelberg. Apply until 12 August https://t.co/JSjiR7nUSc https://t.co/025Pni1DFg
@RuxandraTeslo for loops are just fine in #Rstats, I don’t get that kind of dogmatism. Code read- and maintainability almost always trump speed. Your time is more valuable than that of a computer. The speed of for loops has much improved in #Rstats over the years.
A separate… (1/2)
@RuxandraTeslo issue is that of spaghetti code, to which a for loop construct may or may nor contribute. So, put long and/or recurring expressions into a function, set up the data nicely before the iteration (e.g.: “tidy data”), and the like. The latter often also helps speed.
@RuxandraTeslo for loops can be even more efficient than apply-idioms, e.g. below (less book-keeping, no extra allocation of temporary vectors) https://t.co/IllpTjPLP9
@RuxandraTeslo One real issue with for loops in R is that at each iteration, the interpreter has to assume that all name bindings may have changed, so all lookup of variables, methods etc has to be done each time from scratch. So if possible, move expensive lookup operations outside the loop.
@RuxandraTeslo OTOH, apply-expressions are easier to parallelize, since (afaIk) R can assume that each iteration is independent of all the others and only depends on the state of the world where the *apply is encountered.
@RuxandraTeslo Whenever the R interpreter encounters text that is not a reserved word (like for, function…),it searches through R’s memory for objects with matching name, according to the search path. This is true for normal variables as well as functions. Esp with OOP this can be expensive
@RuxandraTeslo And in the body of a for-loop, this needs to be done over and over again, even if the searches always yield the same result (since, in principle, it might not).
By @K_Imkeller:
The metabolic balance in colorectal cancer is maintained by optimal Wnt signaling
-impact of context-dependent genetic interactions on cellular phenotypes of a central cancer driver -quantitative modulation of oncogenic signaling
Inter-Institutional Postdocs in Heidelberg/Mannheim Exciting opportunities for computational biologists / bioinformaticians in cancer, cardiovascular, neuropsychiatric diseases, genomics, synthetic immunology, molecular engineering, medical technologies https://t.co/NGNEyRuk3z
Research grants for doctoral candidates, postdocs and established researchers affected by the war in Ukraine, for guests stays at Univ. Heidelberg 1-3 months. https://t.co/NAcB56sT6N https://t.co/abq6MOYnb1
Inbox zero! Thanks to August in Europe. After living with a bow wave of hundreds of unread messages constantly no matter how frequently one replies, and the anxiety of the “unknown to-dos”, this is good. https://t.co/ZBa6oLjY2C
Just a picture of Heidelberg, the uni campus, the Rhine valley, the Palatinate on a nice summer evening 🚴♀️ https://t.co/uEpM3UZ0ti
Highly recommended movie. https://t.co/6AAgnSJ3ln
This article is on fire: “The lack of scientific precision and detail in DL publications… At best there are some high-level diagrams. No pseudocode. No equations. No reference to a precise explanation of the model…” https://t.co/CVUxafpK5J
“…sometimes it is not even clear what the inputs and outputs […] of the described model are. Of course someone experienced would usually be able to correctly guess, but this is not a particularly scientific approach.”
“If there is some explanation in the methods section, it is often disconnected from what is described in the experimental section, possibly due to different authors writing the different sections.”
Kudos to Mary Phuong and @mhutter42 for this effort to make DL more accessible, and more scientific!
@nomad421 @mikelove Disagree: 1. Often, the role of the HT experiment is as a screen to generate hypotheses that are followed up mechanistically / causally by other means. Too many replicates for screening are then a misallocation of resources 2. Instead of many replicates of, say, a …(1/2)
@nomad421 @mikelove …drug treatment, a concentration titration with one replicate per concentration will often be more informative. Replication needs also differ dep’ng on whether you’re doing an experiment (well-controlled) or a study (many uncontrolled variables). https://t.co/h1DMMuGhRi (2/2)
@nomad421 @mikelove In an experiment, 2 (or max. 3) replicates are usually enough to make sure it worked, and any additional resources better to go into other experiments, e.g. perturbations, other data types. In a study, e.g., of patient samples, often hundreds are necessary to see the signal.
@IanSudbery @nomad421 @mikelove Yes - continous variables provide quasi-replicates if the effect is smooth
It’s never too early to start writing. Only when you write, you realize what you should have done… https://t.co/yFz0AKKD6b
A reminder that: good journalism costs money, nowhere we can take democracy for granted, paying for content is a contribution to the future of liberal civil society.
Support your local, national and international news”papers” https://t.co/zLojn9EWlo
For those in and around Zurich https://t.co/eeumjqNLG3
Great teamwork by @hollygiles96 and Peter-Martin Bruch dissecting three-way interactions between drugs, cytokines and mutations in primary CLL using combinatorial high-throughput drug screening. https://t.co/BE7WfjKXLr
@tanya_shapiro It was a warm summer evening in 2001, I was toying in Matlab with ideas for analyzing μarray data that I googled to be reinventions of AVAS & ACE in the acepack package. So I installed R 1.2, cursed c() and <-, discovered data.frames, tapply, lm & fell in love. Never looked back.
@mikelove @tanya_shapiro Well it’s the same ‘free’ as in when scientists give away their papers for free.
(In theory, anyway. In practice, we still need to improve the reward system for research outputs.)
@mikelove @tanya_shapiro Yes, some extremely generic, object oriented way of subtracting two sets of 100k numbers from each other that managed to throttle the CPU to making one FLOP every few milliseconds.
@bctallis How?
Summer job offer: Am looking for someone experienced in Rmarkdown and R for a book-conversion project. Should be ca. 40h, work is paid and can be done remote-only. Interested? DM me.
@baruqrodrigues DMs are now open for everyone (previously to followers only). Email contact via: https://t.co/Eyp82Lkj5K – please include a portfolio of previous work examples!
77 years after WWII, SPD (social dem. party) led Germany is soft-pedaling on a fascist regime waging a murderous genocide—against a nation that already had millions killed by the original Nazis. It is bizarre and shameful.
(And how lucky we are that others are more responsible.)
This request is no longer open – we found someone excellent, will be happy to report on the outcome soon. Excited to be migrating the MSMB book into quarto!
@EMBLHeidelberg @embl Preparations for when EMBL expands to being the Global Molecular Biology Laboratory, GMBL.
If you’re using a US keyboard and need to type German Umlauts, this by @borchers is really simple and good: https://t.co/ah5jBi3xtA
Dankeschön!
https://t.co/cL2By7EXjk enables you to search the (up-to-date!) help pages and vignettes of all packages on CRAN (and R itself), and also to link to them, e.g. https://t.co/SNTLhAVpTK
I hadn’t been aware of this, it’s pretty cool.
… the people administering the electricity budget. And you, know, people.
Of course an example how mundane governance issues can have big implications. (2/2)
It seems like many organisations still have long ways to go in becoming more sustainable. Just learned of a case where it took months and many meetings to upgrade a fridge to make it more energy-efficient. Since the people who administer the fridge budget are different from (1/2)
BTW, the upgrade will pay for itself within months and then be profitable.
@dagarfield That’s not what I said.
And it’s also a small, understandable metaphor for much that is happening in the world: externalized costs. Germany’s energy & security policy, everyone’s fossil fuel consumption, big tech’s hijacking public discourse to generate ad revenue, etc
One can teach multi-dimensional scaling using the UScitiesD or eurodist data in R. I added a variation on this theme to the MSMB book: https://t.co/twZIOIuW21 (Fig.9.1-9.7)
🇺🇦 Happy Independence Day 🇺🇦 !
(Some layout features, like figure aspect ratios, sizes, zoom-ability are not yet perfect in the book’s online version, we’re working on a new quarto version.)
Pet peeve genetic interaction mapping: counting them& thinking it’s meaningful. Biological systems are highly interconnected& eventually everything interacts with everything. That’s not a deep insight. What’s of interest are the strong interactions, where “strong” is subjective.
@Dey_Gautam Don’t you also need subjective thresholds for “selective pressure” or “some link to fitness” (or are limited by statistical power concerns that are essentially arbitrary consequences of your study design).
@Dey_Gautam Measurements are (by nature) noisy, and thus statistical power of detection depends on your study design, which implies an essentially arbitrary threshold.
How to infer tumor-specific cancer dependencies by integrating ex vivo drug response assays and drug-protein profiling. Cool work by Alina Batzilla (@alinabatzi), Junyan Lu (@JunyanLu1118) with Thorsten Zenz (@ThorstenZenz) & colleagues. Now in https://t.co/n8rrEUZuyn https://t.co/heCOotQVHQ
This. (Update “TV” to include social media.) https://t.co/znwO9SA6ft
e.g. paper writing, cover letters, responding to reviewers, grants, giving talks, doing interviews, networking at conferences, etc. The little homework of writing a compelling text with the comfort of dictionary, time, and friends who can give feedback is the smallest worry.
One of the stranger arguments against narrative CVs in science (and thus, by implication, pro simple rankings by journal IF or ‘reputation’) is that they create bias against non-native speakers. As if! All professional activities in science depend on the English language…(1/2)
Been googling for references for the idea that causes should precede consequences, and whoa it’s a rabbithole of modern physics and philosophy of free will.
@kiran_r_patil AfaIu: Direction of time, or (ir)reversibility, is not straightforward at all scales of physics (except for thermodynamics, which however, has no inherent time or space scales at all)
… electricity, X-rays, semiconductors, lasers, aviation, mRNA medicines, microscopes
I saw someone ask “What’s the most important problem in science / Why don’t you work on it?”
…and it’s just so not how fundamental science works. We literally wouldn’t have anything if people had always just worked on the next obvious, applied thing.
From prokaryotes to eukaryotes, the basic concept of an enzyme is similar. But there is a lot more compartmentalization, regulation, activators, inhibitors. And I think it’s a beautiful metaphor for the evolution of academic institutions.
Revising a scientific manuscript:
look at all adjectives and either replace by a number or measurement, or remove.
@DrAnneCarpenter @eLife Thank you for sharing this, Anne!
Besides more sanity for everyone and better quality science, smaller lab sizes also means relatively more jobs for group leaders, and an easing of the postdoc - PI career step bottleneck.
@DrAnneCarpenter @eLife There’s a bit of motion in how to do research assessment (e.g. https://t.co/tTVe4dBwN1) and this could be important, as many of the traditional incentives for PIs are to have big group sizes.
Arguments against using “Interestingly,…” in a paper: -if a result contradicts previous views, state that explicitly& provide reference -if not, drop the adverb& get on with reporting the result -you don’t want to imply other results are not interesting, nor use it all the time
@MDLuecken It’s a matter of taste, of course. IMHO flow should come from the logic of the reasoning, almost like in a criminal case, to coolly prove a statement using the available evidence. I care about the subject matter of a paper, not the state of mind of the authors when they wrote it.
Слава Україні.
@PetrovADmitri Interestingly, I think we agree on the objective, just not the means. Of course a paper should have personal style, be engaging, well-written, fun to read. On adverbs, I was thinking of @StephenKing’s good advice in his book “On Writing”. https://t.co/Tivdg7smzl
Second law of organizational thermodynamics: the number of committees is always increasing. (It is much more likely for a new committee being formed than for one being disassembled.)
@jonathancairns There is certainly hot air released.
@shazanfar Welcome to Heidelberg!
The livestream of the Bioconductor conference #eurobioc2022 this week Wed-Fri is now online: https://t.co/F1jDcsDAj7
Conference programme: https://t.co/z6lpYkk1uJ
Unexpected consequence of the drive to save energy: retirement of so many old servers providing still loved compute & web resources, that have been burning away kWs for a decade or more and now need to be migrated to VMs with newer PHP, CentOS and whatever…
@larsjuhljensen Me too 😱😱😱
@bctallis Quality indeed. And you can save electrons with an included electric heating element. https://t.co/VfriPRY1I5
@bctallis Really well. Good coffee and easy to handle. At some point the heating elements gives up, but this is counted in years, thousands of servings. And one needs to be a little careful to wait after washing the base, to avoid short-circuit / triggering residual current protection.
Oh, you serious…? https://t.co/9eQ2cJGaG1
@FredOnion There is e.g. https://t.co/Yof6pBLEex and I think there is also relevant on-going work by authors of the Matrix package and DelayedArray backends that support such matrices via HDF5 – might be a good question for https://t.co/vxGBeMfmyz
@FredOnion Background: R’s built-in integer arithmetic currently goes to 52bit (by “abusing” double prec. floats),e.g.: > 0xfffffffffffff-0xffffffffffffe [1] 1 > 0xffffffffffffff-0xfffffffffffffe [1] 0 but R is extensible so anything is possible in packages and user-defined data structures
@tiberi_simone @Unibo Congratulations, Simone! Great news.
Deleting your old files to save electricity is the new do not print emails to save trees.
@TanyaAneichyk Compared to imaging (light, cryo-EM) this is “tiny”. And afaIk places like EBI are looking into this very keenly. And it now makes even more sense to move data & compute centres to where electricity is sustainable and economic.
Many Thanks to our fantastic participants and teachers! https://t.co/TrvI447nCO
Also thanks to Susan Holmes @SherlockpHolmes for spending the week with us and so many lively discussions on multitable methods and much else with participants and other teachers.
A reminder of the free online version of MSMB: https://t.co/Qjlp7WmCf0 https://t.co/obiejJIy4R https://t.co/TrvI447nCO
Controversial take: seeing a Wilcoxon/Mann-Whitney test in a paper/preprint correlates with no (recently trained) statistician having been involved.
@nomad421 @mikelove There you go. That’s why it said “correlates”, not “1:1 corresponds”. I am sure there are important and well-reasoned counter-examples.
The observation I wanted to share for y’all’s enjoyment is the many instances based on hearsay and tradition.
@timtriche @jmirobla This, and the weird null hypothesis and properties of the test, e.g. “Characterising transitive two-sample tests” https://t.co/dJIUmwRZii
Btw, I’ve nothing against rank transformation, this can be very appropriate, it just seems strange what follows after that in Wilcoxon etc
@jmirobla @timtriche Normal quantile transformation if you must. But in many cases, regular t-test, ANOVA test etc work just fine - normality is a sufficient assumption for easy mathematical proofs, but by no means necessary for good behaviour of these tests in practice.
@NathanClark111 @jmirobla @timtriche You can simulate data (non-normal but same means in the groups) and look at the histogram of resulting p-values. It’s really hard to make it non-uniform. E.g. https://t.co/MXCqkCXEwi
A much bigger issue is deviation from independence (“batch effects”, bad “normalization”, .. 1/2
@NathanClark111 @jmirobla @timtriche confounders). These can easily lead to non-calibrated p-values, i.e. enrichment of small p-values even under the null. 2/2
On the user interfaces for bioinformatics tools discussion: Instead of more GUIs, it’d better to have more exemplary quarto (or equiv.) docs that show analysis paths from begin to end, mixing and matching multiple tools where needed, and that users can adapt to their own data.
@chrisnightwing @jmirobla @timtriche Please check the difference between necessary and sufficient assumptions.
@IanSudbery @jmirobla @timtriche Interesting! It’d be great to be able to explore these examples, a la https://t.co/MXCqkCYclQ or quarto doc. (Btw what do you mean by heteroskedasticity in this context?)
A recurrent theme in the responses is that people long for certainty - if not in the outcome, then in the process. The Wilcoxon makes the least assumptions, so it must always be “correct”. But hypothesis testing is fundamentally about reasoning with uncertainty: making … (1/2)
…rational decisions based on noisy and/or incomplete data. That world is complex, there are no easy answers, there is no certainty, just probabilities. Like everywhere else, be suspicious about easy one-size-fits-all answers. They may be true some times, and often not.
On the way at 300 km/h to an EMBL faculty meeting near Paris. EMBL’s labs are distributed across six sites in Europe, and Paris is centrally located that almost everyone can come by train using the highspeed network. https://t.co/MqMKpot5zj
Sigh, translating a (well-ranked) grant application from English into German for final approval is a thing. Now making sure to use as many genitives, long composite nouns and 1/2-page long sentences as possible.
@MagnusRattray 🤣
@JuliaGustavsen Visualizing big matrices (genes x samples etc.) using heatmaps and PCA. Also, the thing with Excel, gene names and dates.
CZ Software Mentions dataset, a new dataset of software mentions in biomedical papers: https://t.co/JYH8kZOQR4 Tabulation/clustering of ~2.5 million software mentions in paper full texts.
Btw, while this is currently most painful and acute in security & energy, it also extends to other areas…. science and research…
@MagnusRattray @bparsia @Office I’m doing almost everything with Googeldocs (“available offline” mode) nowadays and haven’t missed MS word yet.
I used to use this analogy to motivate importance of data preprocessing& infrastructure in AI. Now, more aware of the bad implications of the fossil fuels business e.g. in Middle East& Russia, not doing that anymore–but wondering whether the analogy still works, in a sinister way https://t.co/p8WafNPRNR
@GonzaParra_ Grant rounds normally have a finite budget, very often the number of good applications is >> money available, and even the most perfect ranking is ‘noisy’ for proposals that are nearly equally good. So acceptance/rejection has random aspects, one should not take them personally.
@GonzaParra_ Very true. Many inefficiencies in the academic funding system that could be improved. Its decentralization is a challenge (but also good in other ways).
In the best case, the process of writing a proposal is also beneficial for the author themself, I hope that’s true for you.
@FredOnion Have a look at the (under development) SparseArray https://t.co/1acUN8fFor which can store sparse arrays of any dimension and atomic data type and >2^31 nonzero elements.
I’m all for questioning authority or interdisciplinary cross-fertilization but phew, the cringe when a techbro discovers a new field and comes up with the most naive solution based on something they saw in grad school, oblivious to many others may have worked on this for years.
(I admit, as a former physicist, of having been guilty of this kind of thing, too, some times. Especially when you enter a new field, it’s impossible to immediately know everything. There’s a trade-off or good balance somewhere.)
A difficult winter is ahead for Ukrainians. Show your solidarity by sending CARE packages. They can be sent at no cost for the sender via DHL, and will be provided to where the needs are most urgent by Ukrposhta. https://t.co/zfvWnlZOFc
@LuciaScience @mikelove @daniela_witten @drisso1893 @stephaniehicks Together with the other replies to this thread, perhaps Figs. 6.2-6.4 in https://t.co/qE6KwDWl0i and Sections 6.7-6.10 in the same chapter can also offer some useful visualizations and arguments. (From the book with @SherlockpHolmes)
Thing #28 to re-learn after the pandemic: the requirements on font and figure sizes for an in-person presentation projected to a wall are different from those of a videolecture to each audience member’s screen.
@IanSudbery I don’t think it’s very controversial that academics have to justify their salary& all the other resources they get. But there is a range in the granularity at which this happens, like from “you’re great here’s your department& tenure for life” to precariousness over every pencil
@IanSudbery … different countries/systems have chosen different spots in that range, some more excessive and counter-productive than others. I do think that being able to take long-term bets and a certain amount of trust are good for both scientific outcomes and academics’ sanity.
@IanSudbery (And of course the overall amount of money in the system is just as important as its allocation.)
@IanSudbery Wolfgang Huber 🇺🇦 Yes, agree 💯% – not argue about the need to make bids for work, but argue about their granularity and exact nature, and perhaps also the overall amount that society is willing to fund.
Probabilistic modelling of transcription dynamics in whole embryos and single cells Magnus Rattray @MagnusRattray ELLIS Life / NCT Data Science Seminar: This Wednesday 11:00 CEST (via zoom) https://t.co/7LbqDZmuxr https://t.co/KHjRIsOG1J
Beta release of a quarto version of “Modern Statistics for Modern Biology” with @SherlockpHolmes
With many thanks to @CrowellHL and @grimbough . https://t.co/qizuuTqGzQ
@geertvangeest @SherlockpHolmes @CrowellHL @grimbough The R code is here: https://t.co/IMWLFfoe8n
@geertvangeest @SherlockpHolmes @CrowellHL @grimbough Thanks. For use in your teaching, I suggest adding a link to the book site to your website. (Hard to see a use case of copying the sources…)
For the followers from Heidelberg: there is some good stuff under #30Ideenfür2030 by @TheresiaBauer Mayoral election is on Sunday 6 Nov and it’s important. Go vote! https://t.co/l7rt1KAkPH
@arjunrajlab Am trying to see it through the glass half full/half empty perspective: be happy about what you get done, not unhappy about what you don’t.
Signal is better than Whatsapp anyway.
@arjunrajlab I do apologize to everyone to whose email I respond too late, I know it’s not good, but sometimes it’s just physically not possible.
@arjunrajlab And of course email is like the Lernaean Hydra, for every email one responds to, two new ones arrive in the inbox.
Congratulations, this is fantastic! https://t.co/fgrpdCYGJ9
Timely and good move updating their Open Science practices by one of the largest science funders in DE. https://t.co/Ido47sN3jd
@DrLachie A segmentation is a model, and “All models are wrong, but some are useful” is one approach: define the quality of your output not w.r.t. intrinsic criteria or comparison to ground-truth, but to its usefulness for the scientific question or downstream users https://t.co/wbC1eC7Z7T
Would you like to host the European Bioconductor Conference 2023, or join the planning committee with the aim of gaining experience to perhaps host the conference in the future? Find more info and get in touch via https://t.co/KyYGymjgFw before 6 Nov.
Multiple sequencing in the evening news via @richardneher https://t.co/91skeAZJht
Stay on Twitter, but join the Resistance. https://t.co/YbqyaXiUXE
@lgatt0 Fully agree with the principled and theoretical reasoning, but in addition there are also simply better, more efficient uses of your time & attention.
Hey world! I just supported The Kyiv Independent on @Patreon, and you can too! https://t.co/wRvTz5tDM3
Congratulations Sascha Dietrich @DietrichLab on his appointment as a full professor and director of the clinic for haematology and oncology in Düsseldorf. Sascha is a fantastic collaboration partner in our joint MMPU group (https://t.co/ajOQ22RMtc). https://t.co/26A8eFQRFQ
Writing evaluation letters for promotions can be a chore esp. when the to-do-list is already overflowing — but it’s also nice to be reminded why you like your colleagues and love your field.
@betowbin @LS2Switzerland @unibern @IZB_unibern Great workshop!
Now… are theoreticians technophobes? I deeply support the push for more theory & conceptual thinking in biology (have PhD in theor.physics myself). Also agree that use of technologies & computers is sometimes excessive. But do these two topics belong together?
Congratulations, @BenLehner ! And wow, what a great catch for the Sanger Institute. https://t.co/2nXP3KZlR9
@bejcal @SherlockpHolmes Thanks @bejcal !
@daniela_oaks @whuber@mastodon.social
@paulpharoah I’m not an economist but I think you need to take into account not only expenses but also incomes. In the current world situation, USD is dearer compared to e.g. GBP because US is where the world wants to put their money. Finally, a progressive goal is not low but fair prices.
@David_J_Adams Standard protocol for me on trips with long jetlag, when I get up at like 3am local time to do a few hours of work in the hotel room (and nothing useful is open anyway)
Go to the mayoral elections if you are an EU resident in Heidelberg https://t.co/pE4EUimj92
@nomad421 In Rust. And you’ll be rich and famous.
херсон ти красивий Слава Україні!
and Europe! https://t.co/tCK9KXxu1t
Here’s a recommendation of the book ‘Science Fictions’ by Stuart Ritchie (@StuartJRitchie, https://t.co/oZaoywQifr) to anyone who expects to navigate the professional science world or is interested in its workings. TBH, when taking the book as a beach read to … (1/4)
It starts with how the replication crisis became evident over the last years, what is is, and dissects the four major flaws that underly it: fraud, bias, negligence, hype. It discusses perverse incentives and, looking forward, ways to fix science, incl open science, … (3/4)
a family holiday in the Canarian sun, I expected no more than ‘worthy and useful for work’. In fact it is fast-paced, full of breath-taking and grotesque examples, and nicely narrated. It has many observations and ideas that will shape science in the 21st century. (2/4)
research assessment reform, team science, preprints, new journal models (spoiler alert: no magic bullet or panacea, but each of them a positive change in any case). (4/4)
Congratulations! https://t.co/KGdozoTLrN
@UniHeidelberg Simon Anders should also be in your list. https://t.co/LqttSsYhGf @s_anders_m https://t.co/mrHfJInIir
@UniHeidelberg Btw, rankings such as this by Clarivate belong in the same category as Impact Factor, h-index, represent a toxic, unhealthy, outdated mode of research quality assessment & I hope also @uniheidelberg will update its practices to be a more state-of-art, e.g. https://t.co/cFgLkF3cyP
@UniHeidelberg (… and I am not saying this b/c sour grapes, https://t.co/0pI1shif9m or https://t.co/iqZqXli254 …)
@EMBL postdoc fellowships are open to researchers with diverse education backgrounds (life sciences, maths, chemistry, physics, engineering, ecology etc)& all nationalities to pursue ambitious interdisciplinary research projects in an international setting https://t.co/1PlNuRI4H1
While Twitter is falling apart… here a video (in German) explaining young Heidelbergers why not to go the mayoral election https://t.co/n0e8s7YrKy
I joined Twitter in 2016 for the science, to get highlights on new papers, talks, tools, and sometimes talk a little about my own. During the pandemic of 2020/21, I enjoyed learning about the newest research and other developments around the world directly from experts in…(1/3)
connect to an entirely different set of experts and politicians in the face of Russia’s brutal war, and I am grateful for the many voices from Ukrainian society I could follow and learn from about their country and fight (Ukraine will win. Democracy and decency will win.) (3/3)
such an immediate manner. It was also great to stay in touch with scientific colleagues everywhere while there were no more conferences and visits; at all levels, from serious science to banter. For the past 9 months, it was enlightening and encouraging to…(2/3)
Postdoc position in phosphatase bioinformatics — in beautiful Freiburg. https://t.co/aMLEnsjwQ8
@MAJAFL Either Maja has joined Twitter 36 years before it existed or the wheels are starting to come off. https://t.co/ydcI2z0PFb
@nomad421 “This is Elon’s platform now” Idk, the concept of ownership is not that simplistic, and different takes on it exist in different legal and societal systems. E.g.if someone buys the mall in the center of the city,in most countries they are still not allowed to just set it on fire.
Don’t be that person in the institute that new applicants are afraid to meet.
@EHunterChristie This is a forever shame for DE, but not really surprising given e.g. Scholz’, Steinmeier’s and others’ actions. Now if they could apply their same cynicism and send Ukraine as many weapons as possible to shorten this war the other way.
Bioinformatician position in pediatric oncology at the Princess Máxima Center in Utrecht—genomics or proteomics data analysis, multi-omics data integration https://t.co/ZBGxLCfze1 https://t.co/ISIdlTRz4n
For the Heidelberg followers: please vote, 8-18h. https://t.co/EIPjWPnx5b
@giorgilab @BardinLab So what is the problem? The work did get out so anyone who cares to read it can do so, and subsequent funding ‘reward’ was apparently commensurate to effort.
@bhklab @_psmirnov Congratulations Petr! Looking forward to working with you from Jan 2023 on machine learning in cancer research with @AurelieErnst in @AIHCluster !
Thank you, Tom, for being such a great colleague! https://t.co/sifEFJhaI5
@daniela_witten More along these lines 🧐 But of course the technology is amazing. https://t.co/n5bsREsG1h
Very good thread about science writing. Sometimes I paraphrase it: “Only when you write it up, you realize what (experiments, analyses, …) you should have done.” https://t.co/mnpf590DcM
Well said.
(https://t.co/Qjlp7Wna4y and https://t.co/obiejJJ5Up with @SherlockpHolmes ) https://t.co/nycvBgp4Wr
Impressive. https://t.co/A1RDoNyGZg
@michaelhoffman After batch correction and normalization, everything is fine. Complicated non-linear batch corrections are the best.
This, from a co-founder of openAI and a developer of chatGPT. Note how these requirements are met by #Rstats capabilities, data wrangling, visualization, tidyverse, shiny, descriptive statistics, … https://t.co/vbFHb4EXFI
@MarcusFaber Guten Tag, Herr Faber, Ihre Partei ist doch in der Bundesregierung, gibt es darüber vielleicht Möglichkeiten, etwas zu machen?
@camille_goemans @EPFL @epflSV Congratulation, Camille! What great news.
This may be handy for your christmas shopping https://t.co/QErDg6l2eS https://t.co/AGkRERS6Ps https://t.co/9Ifx4gu0Nx
@DN_Fitzgerald Well done. Exciting to see your work and its results “coming out”!
@biorxiv_bioinfo For the record, I’m one of the authors of DESeq2, was not involved in this effort, have no idea about its quality, and find the appropriation of the name problematic, intellectually and practically (implementation details can and do matter).
@TanyaAneichyk @biorxiv_bioinfo I’m all for re-use and improvement. I don’t think DESeq2 is the end of history and am happy if people build on top of it, or indeed next to it & eventually supersede it. I also don’t want to be involved in everything, quite the opposite. It’s just the name shadowing. This…(1/2)
@TanyaAneichyk @biorxiv_bioinfo …has the potential to create confusion & friction: if the implementations disagree and users come with support requests, who deals with them? Who documents them? Who takes credit (or blame) for differences in results? As also well-put by Björn: https://t.co/kIRzpopT6d (2/2)
@lpachter @biorxiv_bioinfo Please don’t troll me. I did not speak about misconduct (it is not). I expressed unease about having to deal with confused users and unclear credit & blame. By no means near-identical. E.g., DESeq2 supports full range of multivariate GLMs, PyDESeq2 only does pairwise comparisons.
@TanyaAneichyk @biorxiv_bioinfo This is not specific to DESeq2, it applies to any software. I prefer interoperability: interface to the existing tool & spend your own precious time doing something new. If you do have to reimplement, use a different name and make clear it’s similar, but not feature-identical.
@TanyaAneichyk @biorxiv_bioinfo … and this would not be “stealing” as long as appropriate citations and credits are given in the right places. Just like with many other instances of scientific or technical progress.
@TanyaAneichyk @biorxiv_bioinfo No, this is really not the case 😀 I also do not think R is the end of history, but I still very much love and appreciate R (and the community, the ecosystem) and invest in these.
Twitter might actually be good, and profitable, as a marketplace for selling and finding paid content such as newspaper articles etc linked from the tweets.
Perhaps something for the time after Blofeld manbaby.
@adamgayoso @davisjmcc @minouye271 @biorxiv_bioinfo I’d love to see that. Looking forward!
@BorisMuzc @biorxiv_bioinfo @mikelove No offense taken, and I hope none was caused. I am not concerned about quality here—it could be better, could be worse. More about the idea that PyDESeq2 is a drop-in replacement for DESeq2 (as e.g. your abstract implies)&the potential for confusion and extra work from that.(1/2)
@BorisMuzc @biorxiv_bioinfo @mikelove I think a good way forward would be if the documentation and the published material make clear that PyDESeq2 has reduced functionality (only pairwise comparisons, no multivariate linear models), and where functionality overlaps, it’s intended to be similar, not identical.
@lpachter That’s not what I said and you know it. Troll tactics: twist words in someone’s mouth to stir attention. I care a lot about DESeq2. I am also happy if others try to do even better. It’s a free world & competition is good. This here is about labels that differ from what’s inside.
@ScienceScottT @adamgayoso @davisjmcc @minouye271 @biorxiv_bioinfo As long as the name matches what’s inside, that’s great. Py<X> is fine if <X> is a generic math/scientific concept, if your package wraps the existing tool <Name>, etc If it’s your own take on a problem that well-known existing tool <X> address, then perhaps vary the name a bit.
@ScienceScottT @adamgayoso @davisjmcc @minouye271 @biorxiv_bioinfo And this is not legal or otherwise formal advice. It’s just my personal opinion, and in each specific case, good judgement should be used. There is no global “reservation system” for open source software names (although individual repos usually need one, for practical reasons).
Phew, reacting to provocations by trolls is not normally my thing. Maybe it’s the generally more combative state of the world, or the feeling that this platform is falling apart anyway.
@larsplus This person craves attention by stirring controversy and twisting things, a classic troll. I’m not the first target,& thick-skinned enough. It’s a pity (but sadly not uncommon) that he has such a big platform. I’ve long muted that account, just learned about it through replies 🤯
Welcome 🇪🇪 ! https://t.co/qfryzgFZRL
This looks like a great initiative—data science workshops using R and Python. (Online) participation or access to recording of previous for a small fee that goes to help Ukraine. https://t.co/nSkoqEael3
Data science positions at the German Chancellor’s office.
(The current leadership is bumbling and is repeatedly making fatefully bad decisions, but maybe better information and evidence-based decision processes can do some good in the long run.) https://t.co/XLKTnBrPwI
@Matthew_N_B I recommend the talks listed here https://t.co/REBsoRHbZ4 and the papers under “Neighbour embeddings” by @hippopedoid
@amyhherring Baiersbronn for Black Forest scenery and haute cuisine (e.g. https://t.co/Cc6k3zjf5U ) Lago Maggiore or Lago di Como for Alpine lakes and a whiff of Italy Interlaken & surroundings for Alpine highlights (https://t.co/fgQpEglhFD)
@amyhherring To the East, you can gently & comfortably cycle around pretty Lake Constance in 5-7 days https://t.co/wetGw9505t To the West, there is Alsace (e.g. Colmar, Ribeauvillé) and Burgundy (Dijon, Beaune, …)
The online version of the “Modern Statistics for Modern Biology” book with Susan Holmes https://t.co/obiejJIy4R has been revamped, now rendered with Quarto.
All bug reports, suggestions for improvement etc. are welcome—the book is continuously being updated and rebuilt with current versions of R and packages, this also means that there may be (e.g. layout) issues that we do not immediately or automatically detect.
It is easier to deep learn multimodal spatial single cell data on a Riemann manifold than to make figures with balanced and legible font sizes.
Not a very original take, but White Lotus (I & II) is really quite funny.
Thanks to @CrowellHL and @grimbough for substantial help with the conversion to quarto, from our homemade Sweave-dialect and extensions. And to JJ Allaire @fly_upside_down and colleagues for the powerful, elegant and beautiful quarto system.
@pangenomics Other than ‘printing’ the HTML to PDF—no. Main stumbling block is the page breaks, which in a book with so many ‘floats’ would be tedious and tbh quite pointless to maintain. But you should be able to just download the whole thing with ‘wget’. It is static HTML 5.
@tweet2Rbhadani @learningbioinfo Other than ‘printing’ the HTML to PDF—no. Main stumbling block is the page breaks, which in a book with so many ‘floats’ would be tedious and tbh quite pointless to maintain. But you should be able to just download the whole thing with ‘wget’. It is static HTML 5.
@GHGA_DE @embl Talks slides: https://t.co/8EsYKRhtN7 Source code for them: https://t.co/P7VGE0dVEU
Upcoming Workshops for 🇺🇦
“Color Palette Choice and Customization in R and ggplot2” Thu 26 Jan 2023 17-20h “Python for R users” Thu 16 Feb 18-20h (CET, online)
@DrSethMurray It’s a nice side-effect of using Quarto that the book also renders well on little screens like that of a telephone https://t.co/Jl7rfIr39S
On the biggest issue of our time, who would have thought that Boris Johnson and the Polish PiS party have a better moral compass than three successive German chancellors. But that’s where we are.
The CSAMA 2023 Summerschool “Statistical Data Analysis for Genome Scale Biology” will take place 11-16 June in Brixen/Bressanone — mark your calendars https://t.co/JNLwc2D7W0
@Andrea1Mariani We plan to open the website and a waiting list by Jan or mid-Feb, and the formal registration some weeks later. (The reason for the second timeline is the need for some paperwork for the foundation that handles the budget.)
PS: I’m not arguing that having dependencies has no downsides. It does. But by using an environment such as @Bioconductor, the downsides can be kept at bay, and the upsides can overweigh. Thanks to @mikelove for discussion.
Why having many dependencies in a stable package ecosystem can be good for your computational biology research.
@Malarky67 TBH, I would try to stay away from code that is only on GitHub (and not BioC/CRAN) for teaching, or indeed for anything that should work across time and across multiple people’s computers.
A Gentle and Applied Introduction to Rcpp with @eddelbuettel Thu 9 Feb 18-20 CET, online: https://t.co/58dcxuJg1i #rstats @d_mykhailyshyna https://t.co/aJ8dDM3VRI
So right! Also, it’s not that there is a linear ordering of candidates in which there is a threshold to make, or fail. The qualification landscape is multidimensional, much is about matching people to projects, and to other people. And there are random effects involved. https://t.co/TE75klPxKl
Love it when findings from a screening experiment are validated with a ‘validation screen’.
@David_J_Adams It’s a pretty silly (indeed toxic) measure and should not be considered for anything consequential. Then it also doesn’t matter how you compute it.
@David_J_Adams As an applicant, if you think it helps you, include it. Whatever works. As a reviewer, ignore it. As a policy maker or funder, get your act together and improve your research assessment.
@David_J_Adams The question is so bioinformaticsy 🙂 Trying to ‘normalize’ some data into meaningfulness that are beset by fatal biases, noise, and batch effects.
(And since this is Twitter: the above is said in a jovial, all-of-us-do-this manner)
A whole generation of scientists needs to be retrained for giving presentations in a room, rather than by zoom screen sharing, and use appropriate glyph and font sizes in their slides.
Or maybe I should just get a set of opera glasses.
@pedrobeltrao A real challenge, and I 100% agree. Genuine question: how does one make life scientists follow this advice? The decision to write and submit such a grant is a big commitment and will be mostly driven by individual cost-benefit considerations, not generic ‘community spirit’.
Excited about developing statistical methods and applying them to the latest single-cell & spatial multiomics research in precision oncology? Do a postdoc at @EMBL in the Huber group! Apply until 28 Feb. https://t.co/dRfaUQtUxb https://t.co/7SXU9XrQUE
Cleaning up my inbox with messages from the last weeks, and it is amazing how many professional and commercial organisations are finding it completely normal to ask academics to consult for them, provide contents (recorded talks, teaching material) etc. for free.
@fabian_theis So one could reduce the papers/student ratio or the students/PI ratio or the papers/PI ratio or …🙂
@TheVilchezLab Very much agree! I think the market will take care of it over the next years. Reflects an outdated hierarchical mindset where real academic authority can only flow from a Lehrstuhlinhaber(~Chair). Unis also need to rethink recruitment, which is far too sluggish, and remuneration.
@martinmbauer Does not basically any decent piece of education change worldview? Yet there is also a role for value judgment—which change of worldview is better or more important than others.
Heidelberg’s city council made curbside parking slightly more expensive (130€/a instead of 35€/a). Guess what happened. Bulky waste collection requests are massively up. People are clearing out their garages.
Full professorship for Mathematical Foundations of Machine Learning— in Heidelberg https://t.co/fUfHRvvzGB
@MahesanNiranjan Well it’s more like rent—you rent a sizeable portion of public space (~10 m²) to put your car on it. https://t.co/4JSg27giKe
(Molecular) biology outside the lab:
“The organism and its environment” EMBO | EMBL Symposium, 9-12 May 2023, hybrid Abstract submission till 14 Feb, registration till 28 Mar.
Abstract submission is now open for the BioC2023 conference in Boston, 2-4 August https://t.co/9XIxK0GkBF
Submit an abstract by 19 March for
- Short talk
- Workshop
- Package demo
- Birds-of-a-feather session
- Poster
Submit here https://t.co/JdG9diD7fY https://t.co/6Blv1kVp9P
@anshulkundaje How many months (years) FTE are behind a typical paper?
I completely agree with your premise, but in a total costs view, am not sure the final publication costs move the needle very much. Perhaps it ‘looks’ different to the PI if cost components come from different sources.
@anshulkundaje IMHO, the major place for adjustment to get better efficiency in scientific publishing are peer review processes that cause months of extra work or delays without regard for costs per added value.
@anshulkundaje Again, I fully agree that these costs are a scandal, just…: - one glam journal pub per year per PhD candidate or postdoc is a pretty efficient lab. - as long as we don’t fix research assessment (DORA etc.), then sadly those 5-10k are actually “worth it” for the authors.
@anshulkundaje Yes, lots of economics here: - publishers create an artificial scarcity of slots in the journals to raise the price they can charge for them. - PIs can do ‘conspicuous consumption’ of their and other people’s time to signal the abundance their resources
Nice intro to variance-stabilizing transformations as often used for omics data (log, asinh, acosh et al.) https://t.co/0N78DtjvFK
Zurich Precision Oncology Symposium 29 March 2023 https://t.co/PIe4RnspoD https://t.co/z7zyqKhSbA
I was recently reminded of this: “Let users become developers” - a core idea behind S, R, Bioconductor etc. It’s a main reason for their wide adaption — with the flip side that the R code “out there” has variable degrees of professionality 🙂 From: https://t.co/Ht97P1whr4 https://t.co/mwB1hRYAz3
@Anna_Lena2022 Heidelberg So 25.2. 13:00
https://t.co/3YtEztpgZL (Sonntag, damit keine Überschneidung mit der Demo in der Nachbarstadt Mannheim am Samstag) https://t.co/1rOq4u4SUf
@pwilmarth Making things the same is easy. The challenge is to, at the same time, keep those things different that really are different.
Russian fascism is backward-looking and a death cult. Ukraine is the future. For life and humanity.
They brought a burnt-out russian tank to the RF embassy in Berlin. https://t.co/mrlEGR5pOr
Innovative/interesting repurposing of MOFA https://t.co/27acib84Bh
Registrations are now open for the Bioconductor CSAMA Summerschool “Statistical Data Analysis for Genome-Scale Biology” in Brixen/Bressanone 11-16 June 2023 https://t.co/FfbaOjeXay https://t.co/ciHoJc8Dej
There is an @EMBL-branded (commercial) e-Bike service on campus that makes commuting to meetings downtown or on the hospital campus a lot easier, even pleasant on a gorgeous spring day like today.
It’s only 3km, but 200 altitude difference. https://t.co/C67oTlMqrz
… on using the EBImage package to extract numbers and measurements from images, and demonstrate some follow-up “spatial statistics” concepts.
It’s intro level, so the target audience is not image analysis specialists, but statisticians and scientists who would like to (2/3)
I’m giving an online tutorial on Working with image data in R on Thursday 23 March 15h - 17h CET (Paris/Rome/Berlin) in the “Workshops for Ukraine” series
Images are just arrays, and R is good in handling large arrays. I’ll give an overview (1/3)
extract quantitative data from images and integrate them with other data types. (3/3)
@PavelTomancak @embl Unterer Parkweg https://t.co/lmfdFcIzpl
Estonia is leading the way. Really impressed.
Cluster-free differential expression analysis of multi-condition scRNA-seq data New preprint by @const_ae https://t.co/2ErOCXxnv3
Dividing the cells in discrete clusters or ‘cell types’ may seem intuitive, but often doesn’t reflect biology, and is fraught with lots of…(1/2) https://t.co/Ti3mbEP2bJ
manual intervention, judgement, and back-and-forth. Why not delay this step until after the differential expression analysis (DEA), and do the DEA in a continuous latent space representation of the cell types and states. The manuscript presents a method and an R package. (2/2)
@vnzinchenko @const_ae Totally agree. I guess the motivation and also the basic idea are pretty obvious and intuitive—but implementing this in a way that is elegant & general, and that doesn’t have lots of fudge factors or arbitrary choices, took us on a fun mathematical & theoretical journey.
@avi_dsahu @const_ae @rafalab Really good question. Indeed we started out with Gamma-Poisson sampling integrated into our model, glmPCA-like. All the concepts & tools are there. But then we found that it hardly matters. We wrote this up separately: https://t.co/toiipUjhXQ
@avi_dsahu @const_ae @rafalab So we switched to the current approach, which is computationally a little easier, and seems more generic and more modular.
@avi_dsahu @const_ae @rafalab But I’m sure our current approach & implementation is not the final word, it’d be interesting for someone to see this through with a glmPCA-like observation model for Y.
Wow, this is a game changer. Build websites or documents containing R code that runs directly in the browser, without R installed on the viewer’s computer or a server. https://t.co/BLILRdDIyu
@HellmutAugustin @UniHeidelberg @frauke_melchior Congratulations, @frauke_melchior !
@SameOld_SamOld According to the article linked to from my tweet, that’s exactly the direction of travel, but does not work yet with the current release. One issue is that the R in the browser doesn’t have the toolchain to build pkgs from source (C/C++ etc) & relies on a repo of precompiled pkgs
@notSoJunkDNA All at once?
@dagarfield Yes, exciting times. Happy to hear about your experiences with the two approaches — and possible refinements!
@LuciaScience @dagarfield “All models are wrong, some are useful.” Discrete cell types are a model, simplification of biological reality. As long the model is useful to you, you’re good to use it. But if gradients& more continuous changes in cell type, state are important to your scientific question, …
Well-written and thoughtful article about the Human Cell Atlas by The Economist. https://t.co/Gin7Xn6NLS
Postdoc position in spatial and single-cell computational biology for precision oncology at EMBL
Contribute your skills in data science, statistics, ML to an application of latest technologies to a large clinical collaboration on exciting new drug class
Tomorrow 15:00 CET, 10:00am EDT In this intro-level tutorial I’ll discuss handling images, extracting quantitative information, simple segmentation, object feature extraction, integration of images with ggplot2 and dplyr, optical flow, also some basics of spatial statistics https://t.co/IcDwsPBfWJ
I love ffmpeg.
@antifreezeprot There is: https://t.co/NJErLC3Hx1
@antifreezeprot Thanks to https://t.co/OgUK9MWw0v @opencpu
Work-life balance break before embarking on a work trip. https://t.co/PSffA7DDAl
After dinner conversation at precision oncology conference turns to use of ChatGPT in by patients on their omics data.
EMBL Sabbatical Visitor Fellowships
Together with an EMBL hosting team/group leader, scientists can apply for a sabbatical visit at one of EMBL’s six sites: https://t.co/GwBuWguPqZ https://t.co/690CKQabbF
Never forget. Not again. https://t.co/BkDN9kyYYr
@NamurXa Why would you say that? Whose fees? We care about fairness,diversity&inclusion. And yes financial resources are finite, if extra money is needed it needs to found somewhere, we want to give opportunities to multiple visitors& it’s possible not everyone will be happy all the time.
@michaelhoffman Same here
Great work by @const_ae. At the outset of this project, around the turn of 2020/21, we thought that developing more careful count-based modeling approaches, in the spirit of glmPCA or the acosh-transformation, could improve performance of scRNAseq data analysis,particularly (1/2) https://t.co/Rqd1UkjFzf
in the low-count regime or where cells have very different size factors. This should still be true in some situations. But by&large,we failed to generate broad benefits. Simple log(n+1) followed by linear embedding by SVD/PCA into a few dozen dimensions does remarkably well.(2/2)
Submit your abstract for the European Bioconductor meeting in September in Ghent/Belgium! Deadline this week Friday 14 April https://t.co/0t4kMeTqeo
@MDMorgan_abz @BiotechPedro I make a (semi-serious) distinction between bioinformaticians who start with a method and look for problems (“hammer in search of nails”), and those who start from a problem and then look for solutions—no matter whether it’s just a heatmap or a t-test, or something more complex.
@MDMorgan_abz @BiotechPedro The second type typically seems to be a happier bunch 🙂
ragedonate: https://t.co/QNFkE7yjPb
Most of my supervision boils down to: If a complicated new method applied to a complicated new dataset does not give desired results, don’t try to fix it by throwing in yet another complicated thing. Rather, simplify, take apart, try with toy data, …
@KasperDHansen I think we mean the same, just different words
@JanLause @const_ae Thank you, Jan! I agree it would be more satisfying if one of the more sophisticated methods offered clear benefits, maybe it’s a matter of finding the right use-case/benchmark, maybe of more methods. We did consider the size factor caveat you mention, it’s not as easy as that.
Digital desert Germany. Wanted to tend to my inbox in the main cafeteria of one of its elite unis, in between meetings. Eduroam visible but does not work, no other WLANs, cell phone connection (of course LTE) overwhelmed with lunchtime crowd. Productivity null, mood ridiculous.
@JanLause @hippopedoid @CellTypist Btw, thanks for that paper, it was also your preprint https://t.co/txjvI2n7AZ and your discussion with @satijalab https://t.co/GLr8CZEcaU that nudged us into looking into this topic.
topic was mostly exhausted. Our initial attempts at mathematics- and theory-based methods engineering seemed elegant, but failed to materialize substantial bottomline gains. We wrote all up as a kind of review/opinion piece and wanted to move on. An ambitious reviewer and…(2/2)
FWIW, a background note: it was this preprint by @JanLause @CellTypist and @hippopedoid and ensuing online discussion with @satijalab and C. Hafemeister that nudged us into looking into this topic. Years after scRNA-seq had come out and when I had already thought the…(1/3)
a journal editor asked for a huge extension of the benchmark into something much more systematic and comprehensive. I was unsure, but valiant @const_ae took up the challenge and spent many more months doing that. Thanks to all involved!
If someone is into accumulation of power, resources, influence, titles, then why of all places did they choose academia?
There is also Sayre’s law https://t.co/15Tx5sPzgH
Registration is still open for the Biological Data Science summer school 11-16 June in Brixen/Bressanone. It’s a great opportunity to dive into multi-omics, single cell and spatial analysis, data integration, statistics, R & Bioconductor with top experts in the field. https://t.co/ICkOGjiTHu
PIs: send your new lab members https://t.co/UPfS5bVO0Z
TFW when you keep making the same slide b/c it seems faster than finding it in your previous slide decks.
Statistics (and I am thinking in particular ANOVA) is simultaneously so useful and so poorly taught that smart people keep reinventing it.
@yogeshgoyallab @arjunrajlab You could have a look at the PCA chapter in the Modern Statistics for Modern Biology textbook. There are worked through examples on turtles and decathlon athletes that are simple, non-trivial, and intuitive: https://t.co/ioDUCvk1E9 Theory and concepts in the text above.
I officially declare email bankruptcy.
@ivirshup @Bioconductor @grimbough Thank you, Isaac! Interoperability is key. Here’s a good quote by Robert Gentleman from https://t.co/BsOLcDdqgz Monoliths can be successful for a while for a limited use case. Well-managed collaborative networks can be more competitive, agile, flexible, more sustainable. https://t.co/bE57EjAzBZ
@avi_dsahu @lpachter @const_ae Well, we were similarly surprised. We went into this project with the same prior as you mention, and with the Townes/Irizarry paper in mind. It’s good that the finding stirs controversy, I invite you to do better.
Tomorrow: https://t.co/VerS1zinh5
Several interesting workshops coming up in May & June https://t.co/mjd5YsnVkM
I sometimes get involved in discussions on omics data like “How many clusters are there?” or “What is the best clustering method?” My response tends to be, imagine going to a zoo and being asked to cluster these animals. https://t.co/VzpXuKot2t
Someone might say: 2 clusters! {wolf and lion} vs {kitty and chihuahua} Someone else: 2 clusters! {wolf and chihuahua} vs {lion and kitty} Another one: 4 clusters! (based on multi-omics combination of phylogeny and phenotype) Each of them is right…
Or to put it differently, a clustering is a model, and “all models are wrong, but some of them are useful.”
@Francesco_i0ri0 Yes. But the more pertinent point is that there are multiple features (e.g. weight, height, color, species, fearsomeness, cuddliness,…) that one can weight differently, there is no “right” choice of weighting, and each choice results in different distances between the objects.
@ZaminIqbal @baym @EpicFeil_ @LaurenCowley4 @apreston243 Congratulations, Zam!
willfully chosen by the experimenter, and the other a measurement, then it’s pretty clear to call the independent variable x, the dependent variable y, and to fit y=f(x). It gets more subtle if all variables are observational. (2/5)
Should you regress y=f(x), x=f(y), or just look at the correlation of x and y? Much of biological data science is looking for associations in big datasets, and I keep seeing people being unsure about these options. If one of the variables is an experimental covariate, (1/5)
If there is a reason to think one is temporally, logically or causally before the other, then it is intuitive to call it x and regress y=f(x). If one of them is categorical and the other one continuous, ANOVA is intuitive, which becomes a linear model of y versus x. (3/5)
so this is more about intuitiveness and aesthetics. In R, the three options at the beginning of the thread are written, in the linear case, as lm(y ~ x), lm(x ~ y), and cor(x, y). (5/5)
If the situation is more or less symmetric between the variables, then correlation seems the right option. The other options are not “wrong”. Except for edge cases, the bottom line results (p-values, coefficients) will be usually be more or less equivalent, (4/5)
@gsherloc Hi Gavin, in addition to the other great replies, e.g. by @FredOnion @MagnusRattray and many more, FWIW https://t.co/hs5DLp39Y7 and edgeR user’s guide Section 4.9.
PS These approaches correspond to different probabilistic models, lead to different residuals & in principle to different results. What they have in common is that they deliver inference (“p-values”), and I’m talking about situations with so many observations that …
… the model differences negligibly affect the inference, and it is really mostly about presentation.
@ewanbirney Very important point. Taking into account covariates (potential confounders) helps make the hits more interesting, or more likely to be causal.
@ewanbirney In situations such as you mention, graphical models, which try to construct sparse representations of the joint distribution of several variables, are an option (e.g. https://t.co/m6pVAYVtij )
@ewanbirney @sproul_lab Interesting discussion on this here: https://t.co/n6SeuK3jyx I came across it in statistical mechanics, where you have particles whose dynamics are in principle exactly knowable, but choose to reduce the description to probability distributions parameterized by few variables.
A visit by Paul Nurse, director of Crick Institute, and international education and science ambassador for United 24, to Kyiv https://t.co/g00AktEub7
Mission-driven research generously funded by wealthy foundations is en vogue and offers many opportunities, but also challenges and risks: https://t.co/ID0gSNQ6L4 A critique by its former SAB of the surprise decision to close the Center for Protein Research in Denmark by NNF.
A big fallacy in manuscript drafts is to present the result as a premise or objective at the outset of a section that describes a supposedly exploratory or open-ended analysis.
(Confirmatory work on an already well-stated hypothesis is valuable, but often this is not what we do)
TFW you put a rhetorical question in the comments (margin) of a manuscript draft, to encourage clarification, and the author writes a long, well-formed reply in the comments rather than fixing the text.
@gagneurlab @GHGA_DE Great to see this coming out!
Should you always use the Wilcoxon test as a plug-in replacement for the t-test in omics data analysis, because “it makes fewer assumptions” or “it is more robust”? This claim seems to be widely advocated in the bioinformatics world, (1/7)
It is really hard to break (just try it out using simulations, https://t.co/MXCqkCXEwi). It ’s also easier to extend to include covariates (e.g. batches), or more general experimental designs. (4/7)
it can give small p-values in cases with same or similar medians, if only the distribution shapes differ. These are false detections in terms of the user’s intention.
2.) The t-test is quite robust with regard to non-normality. (3/7)
as came up over the group’s lunch today. I am skeptical and generally prefer the t-test.
1.) The Wilcoxon test is often presented as testing against equality of medians—but in fact its null hypothesis is more complicated, (2/7)
Excerpts:“…the (Wilcoxon) test can be a poor method for comparing means or medians of two populations, unless the two distributions have equal shapes and equal scales” “The t-test is sufficiently robust for use in all likely cases,..” A caveat is that there is generally…(6/7)
The literature on this topic is extensive and I’m not trying to summarize it here, a few entry points include DOIs 10.1002/sim.3561, 10.1016/s0895-4356(00)00264-x, 10.1080/02664769823304. (5/7)
no “ground truth”—tests are often used in omics to prioritize genes and pathways of interests, to be followed up. How useful is such a priorization and how often does the follow-up turn out to be fruitful? That’s an empirical question that’s not amenable to theory or simulations.
@ewanbirney That’s a good point. There’s a bit of that in the quantile-normalization/limma workflow for microarray data, which seems to work well. Rank-based transformations can do funny things to data with different distribution shapes though. There is a trade-off between robustness and…
@ewanbirney quality control—I think sometimes you do not want to be robust, but rather raise a flag bad data quality, throw out offending measurements, and proceed with the “non-robust” method.
@olgavitek Exactly. Also, the t-test is naturally linked to a measure of effect size (difference of means), whereas the output of rank-based tests is less intuitive.
@IanSudbery Maybe, but if you care about ROC, why not state ROC? The proportionality factor between ROC and U depends on sample size and makes U slightly unintuitive (https://t.co/LPYl62qb47)
@dnzmarcio Hmm, when distributions have very different shapes between groups, then one might ask whether direct comparison makes much scientific/conceptual sense. Subgroup analysis, mixture modelling, or some other more detailed modelling then seem more appropriate?
Following up on yesterday’s discussion, here’s @tslumley pointing out another weirdness of rank-based tests (such as Wilcoxon), their non-transitivity, i..e. you can have A>B and B>C but C>A. https://t.co/t0Fq4zJVx2
@tslumley You’re right, this is an additional point. Sorry for the undifferentiated lead-in 🙂
@dnzmarcio Very true, but then you’re in classification world, with measures like (partial) AUC or weighted functions of the confusion matrix (misdiagnosis in one direction might be worse than than in the other), and this is really quite different from omics hypothesis testing.
@a_a_yurchenko88 I cited a few papers in my thread, then also see @tslumley ’s
@michaelhoffman Exactly—once your data are in a place like @EMBLEBI, it is their job and mission to make sure the data stay available for all foreseeable future https://t.co/oKjaCqhdbH
Today @EMBL marks a celebration for Janet Thornton, one of world’s leading researchers in structural bioinformatics, @EMBLEBI director 2001-15, and just an amazing leader, role model, mentor and person.
On the occasion of her retirement from duties @EMBL. https://t.co/HoH7aAKa7j
@d_mykhailyshyna Yikes, apparently me too. Some other account names I tried were not. https://t.co/tZEdCbWv8b
@vitaliikl @d_mykhailyshyna Option 1 is true, but let’s assume it’s not causal. To test Option 3, you could sample some twitter handles (the tool does not need login) & record results.
FWIW I also have few tweets on Ukraine (it’s not the point of this account) & those were mostly humanitarian aid related.
How to help Ukrainians during the flood https://t.co/BFB3IARQEO (tips assembled by @TimothyDSnyder)
Arrived in Brixen for #CSAMA2023 a few days early and taking the 4 year old for a ride to Neustift. https://t.co/BllXa15OMa
The calculations just involve mean vectors and covariance matrices. Higher moments may be whatever they are, but are not used. This is a much looser assumption than normality. One can prove that the methods are mathematically optimal, in some sense, if the data are normal…(2/3)
Another minithread on the normality assumption, often stated and fretted about for methods like linear regression, PCA, t-test, etc. What these methods really assume is that the data are sufficiently described by their first two moments: means and variances. Because all … (1/3)
But this does not mean that they sharply stop working if the data are non-normal. The methods may still be optimal for other types of data, or near-optimal, or just good enough. As long as mean and (co)variance are good summaries. (3/3)
Start of Day 4 at #CSAMA2023 with a primer by @jo_rainer on metabolomics data analysis. And a picture from our hike on Day 3, after a deep dive into sc-RNA-seq and statistical foundations. https://t.co/S3hrDNxeFi
@Kachelmann Ja, manche verorten die Aufklärung im 17., 18. Jahrhundert, dabei ist die Arbeit längst nicht vorbei. Danke für Ihre Beiträge dazu! Die Jahreszahl ist aber 1546.
The Dolomites are great for mountain biking for many reasons, but one of them is tragic. During the First World War, there was heavy fighting in the region between Austria-Hungary and Italy, along largely stationary fronts, at enormous costs of lives. (1/3)
the countries have given up their imperialistic ambitions, and EU and Schengen have softened the borders. Of course, one must think of the current tragedy of the Russian war of aggression in Ukraine (although I don’t think there are useful analogies).
(3/3)
Roads were hewn into the steep mountainsides in high altitudes to bring up canons and supplies. Now, over a hundred years later, those roads are just dilapidated enough to make great cycling. The region is peaceful and wealthy, (2/3)
@hippopedoid @SashaGusevPosts @stetson_thacker I wrote a little thing about small p values: https://t.co/TRfVlFfx3c 🙂
Current implement’ns of R use IEC60559 floating-point (double precision) arithmetic for ‘numeric’. But you can do arbitrary-precision computations eg. with Rmpfr (or Ryacas), maybe that’s what happened here? https://t.co/IidAQKVWQ1
@hippopedoid @SashaGusevPosts @stetson_thacker E.g. the Rmpfr package has quite a few reverse dependencies https://t.co/tlyZYfSL4n (I am not familiar with the paper you cite, and it’s not my area of expertise, so I’m just making a generic comment — technically this seems quite straightforward.)
@hippopedoid @seankenneths FWIW, it’s quite straightforward in R, see e.g. manual page for density and distribution function of the normal distribution, https://t.co/1oZac01QLe, the log.p argument. Similarly for some other distributions. I guess the conversion to decimal mantissa& exponent is evident…
Looking at the out-of-control inbox and the calendar planner full of double- and triple-booked time slots it’s good to remember that nobody is irreplaceable 🙃
@pedrobeltrao I started to see it like the glass half full / half empty metaphor: Happy about what I get done, not unhappy about what I miss.
But FOMO seems seems like such a major condition among academics…
@Alexbateman1 @pedrobeltrao If there were only a way to charge these others for the work they offload on you…
@BenCollinsLab @UCDProteomics All credit to the amazing @JunyanLu1118 now at https://t.co/GfqF472aH2
On assessing the quality of research, DORA and CoARA: https://t.co/S82ILEEE53
Very much honoured by this. EMBO is one of the scientific organisations I respect the most, for its role in building a pan-European research landscape, furthering mobility, personal interactions, training, and careers of junior scientists, based on excellence. https://t.co/48HUlDGOsE
Slides from my talk on Research Assessment at @EMBL’s Lab Day, today
The Biological Data Science Summer School in Uzhhorod, Ukraine! https://t.co/A45hF1AjGV With speakers and teachers from Europe and US, and >60 fantastic students from all over the country. https://t.co/Z41mkvsyk2
Thanks Fyodor @fkondras, Laurent @lgatt0, Guillem, @RodericGuigo & others, incl. the local team, for running a great first week!
After having to be in Heidelberg till today, now looking forward to going and teaching the second week, together with good colleagues and friends.
Maryna Korshevniuk kicking off the second week of https://t.co/A45hF1AjGV with a research lecture on genomic medicine https://t.co/GSMOwIy3pk
Robert Gentleman on regression analysis at @bdssummerschool https://t.co/LVhhr13MK4 https://t.co/iBITd0IKiZ
Veronika Kedlian getting ready for the 3rd research lecture of today https://t.co/aPdEEtA9oP
@ewanbirney Thanks, Ewan. … and mathematicians and computer scientists. E.g., his work keeps coming up here in the context of clustering and image segmentation: https://t.co/ApKDdnu3yr
Impressions from BDS^3
Working away on single-cell RNA-seq analysis The river Uzh Kindergarden playground nearby the Uni building https://t.co/97C6FhUbhl
@gagneurlab It is! Students are really engaged and smart. Some hadn’t had in-person classes for 3 years (or never at all at uni level) — due to pandemic and war. Over the last 2 weeks they’ve been having a change of scenery, and it seems, a good time, academically and in social events.
Thank you to Uzhhorod National University for hosting the Biological Data Science Summer School! The logistics were beautifully organized and just perfect. https://t.co/w5kVQUyKXM
The neighbouring Buffalo Ungvar pub provided excellent meals throughout the course and a nice setting for relaxation over beers and pool games
@vitaliikl @mikelove See also https://t.co/ULv8lO4SBU and Questions 4.18, 4.19. Easy to check your parametrizations by simulating data.
@vitaliikl @mikelove What do you mean by ‘normalise’? In DESeq2, we do not ‘normalise’ counts, but model size factors explicitly (Eqn.(1) in https://t.co/TQfmzRhZ3m ). Having alpha_i be independent of j (and s_ij) is a model assumption. One may argue with it, but that’s what we do.
It’ll be even more so in a country that has incredibly talented young people, but where many potential professors and lecturers have gone into other jobs, or indeed to the nation’s defense. (3/9)
- There is a tremendous need for this sort of event. I knew that this is true for mostly anywhere, based on our experience with the CSAMA summer school that regularly attracts students from all over Europe. (2/9)
(Thread) Earlier this month, some colleagues& I organized a summer school in Ukraine for university students. Here’s some takeaways (#5 the most important): 1.Many students didn’t want to leave, and faculty members were asking “can we do it again?”(yes). It was a lot of fun.(1/9) https://t.co/QqPnfRnejF
and strong research and education. We hope, in our small ways, to help (re)build that. 5.The concept is not restricted to bioinformatics or data science. It is evidently cloneable to other disciplines. Calling computer scientists, mathematicians, physicists, medics, … (5/9)
- For some students, it was nearly the first time they experienced in-person teaching in 3 years — as most instruction had been online first with the pandemic, then the rashist invasion.
- Strong civil society and democracy rest on material and intellectual welfare, (4/9)
- Special and deep thanks to our hosts from Uzhhorod National University, in particular the Dep. of International Relations, Igor Povkhan, Oleksii Kurutsa, Serge Vronsky, Evgenia Fedorivna Hayovich; and to Taras Oleksyk and Walter Wolfsberger for mediating the contact. (8/9)
- A complementary and follow-up step will be mentoring networks between students from underprivileged places and backgrounds, and academics who “made it”. There is scope in rolling this out, at scale, in a sustainable manner. 8.The students we met are wonderful people. (7/9)
- If you want to organize such an event, good contact to a local organiser and host institution is essential. Diaspora Ukrainian academics are an obvious mediator, and many are enthusiastic to help. Include local academics working on the topic. (6/9)
Ukraine is winning. Слава Україні!
(9/9)
References: - https://t.co/LVhhr13MK4 - https://t.co/w5kVQUyKXM
@SantusLuisa Thank you for contributing!
This was really a team effort, with great colleagues, each contributing unique skills and resources: Organisers: https://t.co/VOcTBb8jPI Faculty: https://t.co/wie8htTbAN
Job offer: Head of National Facility for Data Handling and Analysis, at Technopole in Milan. Omics, bioimages, techdev and web: https://t.co/yMNjSg5ZwI
@ewanbirney Thanks,Ewan,for fun dinner conversation and the thoughts about thresholding association tests. I can add some comments. Adaptiveness is indeed one of the main advantages of FDR- over FWER-control. As FDR is defined via the ratio V/R, false discoveries over all discoveries…(1/n)
@ewanbirney the total number of hypotheses tested cancels out, as long as the fraction of non-nulls remains the same. As you say, this can be a rather useful and intuitive assumption in many GWAS settings. However, more on that below. The FDR is also theoretically appealing, as it can..(2/n)
@ewanbirney be viewed from frequentist, Bayesian, and empirical-Bayes viewpoints. John Storey did pioneering here in the early 2000s. As I try to argue in the MSMB book https://t.co/L3MWDRDJ8R, the FDR is the more basic and more useful concept than the p-value, and the one that users…(3/n)
@ewanbirney often care more about. For historical reasons, these concepts are often taught and thought the wrong way around. Finally, scalability. Sometimes, not all hypotheses are created equal. For some, being non-null is a priori more plausible, or we just have better data for them..(4/n)
@ewanbirney Then it makes sense to not just pool them all in a big soup (as e.g. the BH method does), but to stratify them into different groups or along some continuous criterion. That’s the idea behind methods like independent hypothesis weighting, see also https://t.co/2U3TTV7Pbo (5/5).
@ewanbirney It’s like with searching for your lost key under the lamppost first — if you have no idea where it is, it makes sense to start looking where looking is easiest (statistical power is highest) and then move on to the harder places later. https://t.co/X7qqUHiu0R
@ewanbirney There is no double-dipping: under the null, the p-value distribution and that of the allele frequency are statistically independent, i.e. mutually non-informative.
@ewanbirney @mikelove a) The ‘cheating’ concern is basically addressed here: https://t.co/9SO1OiQ6dL - independence between filter criterion and p-value under the null. Others have shown how to relax this& use dependent criteria, and explicitly model these. But for our purposes, independence is fine.
@ewanbirney @mikelove The IHW algorithm looks at the aggregate of prior probability (selection argument: low freq is better) and detection power (high freq is better) by optimizing for number of discoveries. So it can combine these different influence factors. However, the current implementation…
@ewanbirney @mikelove rewards all discoveries the same, whereas, if I understand correctly, you like some more than others. This would need a modified optimisation objective. I think it’s straightforward and we discuss it in the JRSSB paper, but it isn’t implemented. @nikosIgnatiadis
@hippopedoid @adp_diaz FWIW, we (i.e., really: Susan Holmes) use these data here: https://t.co/VPbEEGBPGa and throughout the chapter
@hippopedoid @adp_diaz Pretty sure she would! She’s here now: https://t.co/5rdGZ70AK9
Bioconductor Awards 2023 Really happy for all four of them. Highly deserved https://t.co/kEfmIzuJ8k
@gwcarter The main advantage of the narrative ‘research outputs’ lists over a traditional ‘publication list’ is the possibility to add (and put in context) outputs other that papers, in particular, software, datasets, or ideas and concepts that transcend individual papers.
@gwcarter The problem is real, but don’t you think it is even more substantial with glamour publishing?
@gwcarter The time and effort concern is legitimate, but e.g. hiring decisions are pretty momentous (for both sides), so updating 1/2 page of text seems like one of the least places for misallocation of effort…
@Schwarz_MdB @MiRo_SPD Danke.
@nomad421 It’s quite an art form and a livelihood to smell ideas that are in the air, rush to glampublish something with flimsy data, and then let others clean it up with more careful, more tedious work, in “lower tier” journals.
@AOri_lab @genentech @LeibnizFLI @dariovalenzano_ @MariaErmolaev13 @DomenicoFraia @AntonioMarinoMB @AmitKusahu1598 @blue_ceil @GebertNadja Cool. Congratulations!
@arjunrajlab The economics are different. E.g. fixed vs marginal costs.
@arjunrajlab I.e. it’s not necessarily about “idealism”, but different funding and incentives models.
@arjunrajlab Costs for a software product are development, maintenance and support. The first two are fixed, the last probably sublinear in number of users. Cost of entry is low. For a physical product, in addition you need industrial production, storage, shipping. Cost of entry is higher.
@arjunrajlab Funding agencies seem ready for the former, less for the latter. Also, customers may prefer integrating an open source software in their workflow since, if the producer shuts down or discontinues, they have the option to self-maintain and keep going. Whereas there is usually no..
@arjunrajlab such option for a commercial product with all sorts of IP, trade secrets and barriers of entry to it.
These are just some random thoughts, I am not an expert.
How can scientists around the world help scientists in countries affected by war and hardship? Great paper by Serghei Mangul,Taras Oleksyk & colleagues https://t.co/Ym2d59vjzb https://t.co/WcAKeAQ4mN
👇
(and, where applicable, methods / software scripts) https://t.co/bfXwcIL1Wt
Can’t avoid thinking of a workplace organization as a regulatory system (like a gene network) and assigning activator and inhibitor interactions….
I learned from some University travel & accommodation guidelines the ‘principles of efficiency and economy’ and I think that is beautiful. Now let’s apply it to committee meetings.
@JustinMCrocker Yes! In that context it’s remarkable that genomes of “higher” organisms have an ever higher ratio of regulators vs enzymes.
Research Day of the Molecular Medicine Partnership Unit (MMPU) of Heidelberg Univ. Hospital and @EMBL, on Thu 14 Sep 13:00-17:00. https://t.co/sjEB5y76Bh
Apply for a PhD position at @EMBL https://t.co/gfCc1UGSuY
EMBL-EBI is looking for a service-oriented team leader to lead the work on its databases and resources on functional genomics and single cell resolution data (e.g. scRNA-Seq). This is a world-wide unique job with potential for huge impact. https://t.co/wy1RVgnCSt https://t.co/rc3SJAnVcz
Working conditions are great, and Cambridge is lovely.
@arghya_dutta_ @CambridgeUP Thank you, glad to hear it’s useful!
@mikelove The eternal question: intent or incompetence?
@arjunrajlab They’re very distinct. The term “high impact paper” exposes sloppy thinking; not sth you want in a scientist. 1.If they mean “paper in high IF journal”, the fallacy is that IF is an average for a journal, not for an individual paper 2.If they mean “highly cited” why not say that.
@clhubes Yes, I agree so much. Love our Thule Urban Glide 2. https://t.co/us9PnZsvKC
@jordivangestel @ERC_Research @embl Congratulations!
Shop presents Made With Bravery for your favorite upcoming holiday (Christmas etc.) https://t.co/QErDg6kupk https://t.co/85Ls9mRup0
As the standards of this platform continue to descend due to the wannabe-Bond-villain manbaby, I invite you all to follow me on the place where the skies are blue @ wkhuber
@jessenleon Yes, but that’s growing exponentially, doubling time ~1 week or so, afaIct.
@AlejoFraticelli I lost trust in X’s feed algorithm, which is everything. I perceive increasing down-weighting of legitimate topics (incl mine) and boosting of conspiracy & hate. Also, to the extent that my engagement generates value (revenue) for X and its manbaby owner, I don’t want it.
Beautiful picture of Scotland. Without clouds 🙂 https://t.co/uN49oWhC5R
@matloff @stephenjwild Thank you, Norm!
Susan is here: https://t.co/qTo82GtOsp The book: https://t.co/obiejJIy4R … and me: https://t.co/xZkQoYMSTK
Congratulations @Michael_Boutros ! https://t.co/0fEE5SaA5M
@ewanbirney The underlying problem is using discrete categories for a continuum. You want to estimate how well your trained ML model generalizes to data that was not used to develop it, but that other data can be “other” in an infinite number of ways.
Today 16:00, Bioquant Heidelberg: Robert Gentleman on Computational Biomedicine- How will generative AI research, practice and teaching of medicine? https://t.co/D7HGjQzIuc
Ben Hodges and Anton Hofreiter: https://t.co/9113VMPP9S
EMBL Distinguished Visitor Lecture:
Anne Carpenter (Broad Institute)
EMBL Heidelberg, Large Operon, Thu 26 Oct 2023, 13:00
Insights from images: microscopy data for functional genomics and drug discovery
@kieranrcampbell @mo_lotfollahi @david_sontag @RBoiarsky An interesting circularity of categorization here.
@baym Yes. I think it’s partly the shortness of election cycles and (science-)political careers that want quick results, and partly what you say. It’s not a new phenomenon: in 1878, Max Planck’s university advisor told him that “almost everything is already discovered” in physics (1/2)
@baym and advised him to work on something else. Planck then of course went on to discover quanta and laid the foundation for quantum physics….
@LuciaScience Yes. Much of what needs to be said about t-SNE has been said here: https://t.co/dF6J8s3zXZ Fig.1
EMBO|EMBL Symposium
AI and Biology
12-15 March 2024 in Heidelberg with an exciting line-up of invited speakers and interactive sessions. Submit your contributed talk before 5 Dec.
Visualization of package dependencies in R, here e.g. DESeq2. My takeaways: -they can be enormous -the spectacular collaborativity of the R& BioC communities. Developers can reliably reuse inputs from 10s/100s of other projects, distributed around the world& across disciplines. https://t.co/nf8CckOih6 https://t.co/g1S5x4w5eE
And the dependencies and the collaborativity is recursive, which means we can really stand on the shoulders of giants.
@anshulkundaje @BioMickWatson @cshperspectives @mbeisen Very much agree. Maybe also: Professional reviewers like for books, movies. Festivals like Sundance, Cannes, Frankfurt Book Fair…, which can be done by conferences.
@jokergoo_gu I wonder whether this can also be done for Python packages that are widely used in bioinformatics—and if such analyses provide information about (different) development models or community interaction patterns?
@mikelove Similar—I still show up here to read posts of people I respect and occasionally engage with replies. My own content goes preferentially to Bluesky. I block anyone who puts an ad in my timeline. The guy who owns this place is poisonous, and has poisoned it.
@ltronneberg @ManuelaZucknick Additive in the same individuals, or at the population level (i.e. if drug A does not work in one person, drug B will, and overall more people are cured)?
Also, therapeutic window matters—does combination A+B have fewer side effects than 2*A ?
DHL provides free shipping of CARE packages to Ukraine. Winter is coming, and durable food, medicines, hygiene products are needed. https://t.co/ZaVsDtLBSH
Ascona Workshop, 8-13 Sep 2024:
Spatial and Temporal Statistical Modeling in Molecular Biology
Statistical, computational and ML methods & applications to spatial biological data, from spatial omics of tissues to ecosystems and planetary-scale biology
@anshulkundaje Impossibility claims are hard to prove (“Something can be done with Method A but not B”). Also rarely pertinent. More interesting is whether something is easier with A vs B.
Also, “more data is better” or “biological systems operate in space& time” seem not like big surprises.
@pedrobeltrao @GonzaParra_ I’m as skeptical of hypes as anyone, but I think there is something there that won’t dissipate in a few years. Perhaps like the invention of the steam engine or electric grid. People had only vague ideas what to do with them early on,and it took time, but they changed everything.
Giving Tuesday Appeal: Bioinformatics for Ukraine
Empowering Ukraine’s Bioinformatics community through education and collaboration https://t.co/xoNpNNh3KN https://t.co/ZoeE7Quy07 https://t.co/5eutqmNzg0
X’s new business model: pay us or we show your ads on X https://t.co/8pDUMuV0NF
@ItaiYanai @Nature That face-to-face meetings are a good thing has been known before. I am not sure what exactly this study adds: 1. The reported effect size is quite small (perhaps because the D-score is not a very good measure of true innovation)….(1/n)
@mo_lotfollahi
@ItaiYanai @Nature @mo_lotfollahi 2. It does not disentangle correlation from causation (perhaps remote teams and local teams are set up upfront with different types of goals upfront) 3. It looks at output per paper but not at output per invested resources: remote teams may be… (2/n)
@ItaiYanai @Nature @mo_lotfollahi more affordable or more practical than trying to hire everyone into the same place. Output per resource may still be higher for remote teams! 4. It’s a false dichotomy. Many teams meet combine some in-person meetings with substantial remote work. 5. Remote teams can be… (3/n)
@ItaiYanai @Nature @mo_lotfollahi …more inclusive, geographically, socio-economically, or for reconciling work and family care responsibilities. It’s hard to argue that more diversity should not benefit innovation. … (4/4)
@jenseisert @Apple I now largely switched from Keynote to Google Slides. So much easier to collaborate and share. Have not yet been missing functionality.
(For functionality incl. reusability, maintenance and fun factor, nothing beats https://t.co/2Cj2wzOASI 😎)
@falexwolf @LukasHeumos @scverse_team Yes - thinking of interoperable data structures as objects in memory (whether in R, Python, or other language) and not just as serialized files/connections (as in Unix) is liberating 😎
@weinberz @cshperspectives Academic jobs are becoming a buyer’s market.
@IgorUlitsky @cshperspectives “Today it is bad, and day by day it will get worse―until at last the worst of all arrives.” ― Arthur Schopenhauer
Remarkable. Plötner and Yermak. https://t.co/aeBvkUZnX9
Safe Skies, by @TimothyDSnyder https://t.co/XMlhiVpQM7
Donate to help launching a Ukrainian PhD program in #Bioinformatics mentored by senior scientists from around the world. https://t.co/7Z5vxVeBBL
@STOPlabPI Done - https://t.co/bqXxsCW9Zr
I stopped using this account for science months ago – please find me on https://t.co/xZkQoYMSTK DM me for some invite codes (as long they last).
Am still using this account to get news from Ukraine or retweet occasionally.
@arjunrajlab Agree, but would also say that if your data are not worth sharing, then the paper and the conclusions are likely not worth reading.
@bhaibeka @arjunrajlab Science Fiction 🙂
@arjunrajlab I see your point and of course such papers exist. As for your question: human genome, ENCODE, TCGA, ICGC, Alphafold,IPCC, GWASes, … Why are many scientists fine to jump through absurd hoops to get a PDF into a glam journal but whine about the extra work for data sharing?
@arjunrajlab @anshulkundaje @vsbuffalo Are you applying the same logic to your manuscripts (as a ‘science generator’).
@arjunrajlab @jmw86069 @lpachter FWIW my summary on data sharing. It is hard, should be made easier for producers and consumers, co-determines impact, and probably we all need to plan it better. https://t.co/2dHou5Lw9v https://t.co/1I0qJUemsr
@arjunrajlab @jmw86069 @lpachter I agree, and note that I am not arguing based on ideals. IMO it’s possible and legit to publish data-based papers without data or methods papers without software - but then just don’t expect as much impact, visibility or glamour for it.
@alexisjbattle @JHUBME @JohnsHopkins @HopkinsEngineer Congratulations, Alexis!
Ascona Workshop: Spatial and temporal statistical modeling in molecular biology
8-13 Sep 2024 in Ascona, CH
Register and submit your talk now https://t.co/uvIyI0JEXC https://t.co/3J4Q4O1p2g
@BrittaVelten @dfg_public That’s fantastic news. Congratulations, Britta!!
This Elsevier/Ioannidis “Top 2% Scientists” list is a piece of work. According to it, I wrote my first paper a year before I was born. https://t.co/uebqB3B2Hl https://t.co/zSbcSHdhIi
@gsherloc @lpachter Thanks for the accolades, but as the links below highlight, others are even more impressive. E.g.,Tom Blundell published 89 years before he was born, Lord Kelvin 104 years after he died, A.Einstein as recently as 2021.
https://t.co/B9cd89IKVC Spreadsheet https://t.co/bycSvOMhv2
Heidelberg 24.2.2024 україна переможе зло зазнає поразки https://t.co/RA2igicMrh
@STOPlabPI Plus perhaps a better gender ratio?
@STOPlabPI @CantoneIrene @KellerValsecchi Yes, I agree, it’s a process, and I understand from your comments that the current, transparent, public process and the international affiliations are impressive, and good steps in a right direction compared to what was done before.
Panel discussion: From algorithms to biology, with Emma Lundberg, Caroline Uhler, Julia Mahamid, Trey Ideker and Ewan Birney at the EMBO | EMBL Symposium “AI and biology” https://t.co/4IpoyeWKTk https://t.co/RF9agsGswR
Join us for CSAMA 2024 Biological Data Science summer school in Brixen/Bressanone 23-28 June. https://t.co/vq0yYUMVs4
Exciting line-up of lecturers incl. Davide Risso, Mike Love, Charlotte Soneson, Robert Gentleman, Vince Carey, Chiara Romualdi, Ilaria Billato https://t.co/Ogr0zanckv
The European Bioconductor Conference 2024 will take place in Oxford, UK, on 2-6 September.
Abstract submission deadline: 26 April!
Bioconductor Conference 2024 24-26 July, Grand Rapids, MI
Call for abstracts deadline: 25 March!
Open Position: Scientific Lab and Project Manager at EMBL Heidelberg https://t.co/WEZAGm7sbQ
Congratulations, Britta!! This is great. @BrittaVelten https://t.co/rkumFzkIrH
@TraverHart paraphrased in https://t.co/YphI3oSflC
Batch “correction” can (and often will) introduce its own artefacts.
In addition, it tend to distract users from doing quality control—making sure all the data are valid, usable and optimally preprocessed in the first place.
The name “correction” is mostly wishful thinking. 👇 https://t.co/AexfJrABlr
@vitaliikl I mean the name, not the methods per se. “Correction” implying that all is correct afterwards. I prefer “adjustment”, which is more neutral, and implies that one still needs to check whether it actually did make things better. I agree that can happen, but it’s not automatic.
@_canergen Can you elaborate? As a scientist, I would be pretty worried if anything of importance in my results depended on the choice of random seed.
As Frank says, many good points.
Personally, I’m happy how working in academia turned out, thinking that enough of the stuff may actually be useful and definitely is fun. But there was a lot of luck involved. Including some great role models and mentors. https://t.co/KTGphmFnM5
@f2harrell @AmyntasAngelos Fully agree. It’s a jungle. Many wrong incentives.
What gives me a little hope for the future is the broad momentum for open science and research assessment reform (such as DORA, https://t.co/cFgLkF2EJh), which comes both from grassroots, and the top (big funders).
@f2harrell @AmyntasAngelos It’s a genuinely hard problem to distinguish good research from bad, and to decide who gets the resources, jobs, promotions. Research performing institutions who does this more carefully than others have a competitive advantage. On the long run. I hope.
Register before 22 April:
Ascona Workshop 8-13 Sep 2024 Spatial and Temporal Statistical Modeling in Molecular Biology
Statistical, computational and ML methods & applications to spatial biological data, from cells, tissues to planetary-scale
@platten_michael @ERC_Research @Platten_lab @DKFZ @Neuro_MA @unimedizinma @UniHeidelberg Congratulations!
@anshulkundaje It’s also a serious candidate for Goodhart’s law https://t.co/CcHxKTljQe
@larsjuhljensen I don’t know whether your question has a good normative answer. But I decided, given the amount of review requests and other things to do with my life, to disengage from gatekeeping and just review work that I expect to like and for which I want to help improve its presentation.
@larsjuhljensen i.e. https://t.co/gS54ZLiirQ applied to academic publishing
The resilience of Ukrainian scientists
Cell Systems asked Ukrainian scientists how they have been able to persist since the full-scale invasion of their country by Ruzzia. Incl a piece by Svitlana Dekina from @EMBLHeidelberg
Last chance to apply for participation – pre-registration deadline 5 May https://t.co/7uv0eIyXBf https://t.co/2dgb2oO0UZ
@sp_monte_carlo All true. K is a.k.a. as the adjacency matrix and you’re in linear algebra world. IMHO the usefulness of graph models, if the underlying reality is continuous, is sparseness: easier computations, perhaps more interpretability.
One of the best jobs in the world 😎 https://t.co/9mMTowjLXV
CSAMA one-week summer school biological data science, in Brixen/Bressanone, South Tyrol, 23-28 June. Register now for one of the last places. Topics include single cell omics, statistical foundations, R/Bioconductor tools, mass spec, interactive workflows https://t.co/vq0yYUMVs4 https://t.co/QRquAsEWHw
Guess which of these two directions scientists and the funding agencies will be pushing for?
We need more appreciation of infrastructure, plumbing, automation of mundane tasks. https://t.co/g6cfalkirT
Goosebumps https://t.co/CdhM0aoZEz
100% this.
I was initially a bit skeptical whether ‘reputable’ is the best adjective here, but it probably is. Also, I think Yann gives a necessary, not sufficient condition. https://t.co/Ej2Ce6jKds
PhD position - use single cell technologies to understand immune responses to allogeneous haematopoetic stem cell transplantation, with the great Tobias Wertheimer in Freiburg https://t.co/ng9x9YQnkR
Arriving in Ukraine, Uzhhorod for Ukrainian Biological Data Science Summer School. https://t.co/A45hF1ARwt
@larsjuhljensen just meeting the students, so many smart and ambitious people!
@dana_peer Ukrainians are the heroes.
The idea, running summer schools for undergrads, in the country, is very scalable and transferable to other fields. I encourage all the copy this.
Here one can donate for the childrens’ hospital the Russians bombed today: https://t.co/mwyQzoR9aD
The Ukrainian Biological Data Science Summer School 2024 — a big thanks to all the faculty and TAs and fantastic group of students from across the country! https://t.co/gY6b7Hr3X4 https://t.co/RNkQMUVEfu
Independent Group Leader position in Machine Learning (“AI”) at EMBL in Heidelberg, in the broad area of ’omics.
This is an excellent opportunity to transition into scientific independence, with great colleagues in a truly interdisciplinary environment.
We hire based on potential, not achievement. Applicants with industry or academia experience are welcome. Interests in natural sciences, statistics, open science are a plus.
Added bonus: great childcare (kindergarden) from 3 months to 6 years, and a beautiful commute through the forest. https://t.co/eV8tqjMzeH
@ylecun Agree. Sadly it seems that some groups that are not obvsly far right/left, but extremely well-resourced, decided that liberal democracy is not in their interest & that they’d like to try autocracy: tech bros, billionaires, evangelical Christians. Big roles of Meta,X etal. in this
Was reading an intricate ‘data analysis plan’ for a grant proposal, with all the right words like AI, deep learning, cloud etc. but couldn’t help thinking about Mike Tyson’s “Everyone has a plan till they get punched in the mouth” or von Moltke’s “No plan survives … (1/2)
…the first enemy contact”. So, IMHO, more important than a detailed plan is to have the right people, skills, and resources in place and to be able to (re)act quickly upon the data that get produced—if, when, how, and how much—and to rapidly changing scientific questions.
The perfect stoic attitude https://t.co/qdc1x5AP08
100% this. Journal brand (or equiv) can be useful, but for quality standards and trust, not pomposity. https://t.co/dClBLKdJ1c
@TraverHart To be fair, I’ve experienced some good (quality-oriented) review processes at ‘fancy’ journals. In principle, they can pay the editors better and reviewers who are competent as well as busy may be more likely to accept the task.
Job at EMBL Heidelberg: Bioinformatician
Scientific Data Manager and Research Software Engineer in Precision Oncology and Multiomics
@arjunrajlab I agree (in principle 😎), and it’s e.g. how Susan Holmes and I designed the MSMB book. But how do you argue that some examples are good/correct and others are bad/wrong if not referring back to principles, i.e. some underlying theory, abstractions?
@MiRo_SPD @DLF Danke.
@wc_ratcliff I just use this site to read from Ukrainians I follow and other analysts. B/c they’re here.
For science, and my own posts, Bluesky.
@lawrennd It’s a great book!
I learned a lot from reading it. Kudos Neil for this impressive achievement.
@nomad421 Given how important identification of good work is for conduction and management of science, it’s stunning how lazy-operationally and intellectually- many scientists want to be about it.
“Let’s just outsource it to an opaque commerce with overworked editors and unpaid reviewers”
Recommended https://t.co/L9lyenWxdw
UNITED24, the official fundraising platform of Ukraine https://t.co/KDpbJON1zB
Scratching my head… As Dmitry (@hippopedoid) rightly points out, the UMAP objective function is isometric (E(n), rotation and translation invariant; a point often made by Dmitry Kobak). So there are no particular axes.
But there is a space—how to best represent that visually? https://t.co/5wFqQ2cr9D
@hippopedoid I like emphasizing the 2D Euclidean space aspect. But the UMAP objective is also equivariant to scale transformations, and the scale e.g. of log counts is arbitrary, so what does the “10” actually mean?
@NimwegenLab @hippopedoid Yes, it’s a metric (Euclidean) space, this should be shown.
Align with PCA axes? I don’t think so, these maps are exactly not intended to preserve global structure. S.a. https://t.co/wuevCE4Kfz by @hippopedoid
@NimwegenLab @hippopedoid “All embeddings are wrong, but some are useful.” Usefulness is now apparently an empirical fact in the literature. And IMHO the deficiencies can be dealt with.
@NimwegenLab @hippopedoid Ouch. Homeopathy is bad. Please met me explain why I think it’s not the right analogy, and UMAPs etc are not “evidence”.
They are an exploratory tool, first line visualization and hypothesis generation, which need to be followed up by more rigorous methods and validations. 1/2
@NimwegenLab @hippopedoid In your analogy, they are the light-hearted chit-chat a doctor may do during anamnesis, while trying to figure out a patient. Then to be followed up by more serious diagnostics, and well-tested therapies. 2/2
@MarcusFaber Guten Tag, Herr Faber, danke. Das ist völlig richtig, aber auch schon lange bekannt. Was ist zu tun?
@ewanbirney @anshulkundaje Yes to “1st class”. Benchmarking needs a reputation boost. It can be as conceptually and theoretically challenging as method development. Defining what the question is can be the greater intellectual effort than finding its answer.
@ewanbirney @anshulkundaje It’s no coincidence that some of the biggest successes in ML were in ‘closed universes’ (e.g., playing Chess or Go) or in fields with very clear benchmarks (CASP / protein sequence->structure)
Reading the term “Neofeudalism” for the first time and what a fitting one it is. https://t.co/5cCBW3SXLv
@fetzert Analogously in the natural sciences with “novelty” — creates bad incentives and poor results. But what’s the alternative? We do need criteria for allocation of resources. Seems we need clever economists to help fix the system.
Of course it’s a “he”. https://t.co/qyJdVCKi2c
What if the AI has already infiltrated the communication channels among Nobel committee members and just decided to give the price to itself.
And since this is the internet, note that was meant as a joke, and that I think: - Recent progress in ML research is fantastic. - Nobel prices are an anachronism and reflect an outdated, inefficient if not toxic model of science.
Bluesky works.
@MykhailoRohoza I stand with Ukraine. 🇺🇦